Tutorial: Gaussian mixture model and the EM algorithm
MAP573 team
11/10/2020
library(mixtools)Gaussian Mixture Models
We consider a collection of random variables \((X_1, \dots, X_n)\) associated with \(n\) individuals drawn from \(Q\) populations. The label of each individual describes the population (or class) to which it belongs and is unobserved. The \(Q\) classes have a priori distribution \({\boldsymbol \alpha} = (\alpha_1,\dots,\alpha_Q)\) with \(\alpha_q = \mathbb{P}(Z_i = q)\). In other word, the latent variable \(Z_i \in \{1,..,Q\}\) indicating the label follow a multinomial distribution \(Z_i \sim \mathcal{M}(1,\boldsymbol\alpha)\), such as \(\sum_{q=1}^Q \alpha_q = 1\).
The distribution of \(X_i\) conditional on the label of \(i\) is assumed to be a univariate gaussian distribution with unknown parameters, that is, \(X_i | Z_i = q \sim \mathcal{N}(\mu_q,\sigma^2_q)\).
We further introduce the additional notation \(Z_{iq} = \mathbb 1_{\{Z_i = q\}}\) and \(\tau_{iq}= \mathbb{P}(Z_{iq}=1|X_i)\) for posterior probabilities.
We denote the vector of parameters to be estimated by \(\mathbf{\mu} = (\mu_1,\dots,\mu_Q)\), \(\mathbf{\sigma^2} = (\sigma^2_1,\dots,\sigma^2_Q)\), \(\boldsymbol\tau = (\tau_{iq, i=1,\dots,n; q=1,\dots Q})\).
Questions
Likelihood. Write the model complete-data loglikelihood.
E-step. For fixed values of \(\hat{\mu}_q, \hat{\sigma}_{q}^{2}\) and \(\hat\alpha_q\), give the expression of the estimates of the posterior probabilities \(\tau_{iq}= \mathbb{P}(Z_{iq}=1|X_i)\).
M-step. For fixed values of \(\hat{\tau}_{iq}\), show that the maximization step leads to the following estimator for the model parameters: \[ \hat{\alpha}_q = \frac 1n \sum_{i=1}^n \hat{\tau}_{iq}, \:\:\:\:\: \hat{\mu}_q = \frac{\sum_i \hat{\tau}_{iq} x_i}{\sum_i \hat{\tau}_{iq}}, \:\:\:\:\: \hat{\sigma}_q^2 = \frac{\sum_i \hat{\tau}_{iq} (x_i - \hat{\mu}_q)^2}{\sum_i \hat{\tau}_{iq}}. \]
Implementation. Complete the following code.
get_cloglik <- function(X, Z, theta) {
#returns the complete model loglikelihood
n <- length(X); Q <- ncol(Z)
alpha <- theta$alpha; mu <- theta$mu; sigma <- theta$sigma
Xs <- # to write
res <- # to write
res
}
M_step <- function(X, tau) {
#maximization step
n <- length(X); Q <- ncol(tau)
alpha <- # to write
mu <- # to write
sigma <- # to write
list(alpha = alpha, mu = mu, sigma = sigma)
}
E_step <- function(X, theta) {
#expectation step
probs <- # to write
likelihoods <- rowSums(probs)
list(tau = probs / likelihoods, loglik = sum(log(likelihoods)))
}EM_mixture <- function(X, Q,
init.cl = base::sample(1:Q, n, rep=TRUE), max.iter=100, eps=1e-5) {
n <- length(X); tau <- matrix(0, n, Q); tau[cbind(1:n, init.cl)] <- 1
loglik <- vector("numeric", max.iter)
Eloglik <- vector("numeric", max.iter)
iter <- 0; cond <- FALSE
while (!cond) {
iter <- iter + 1
## M step
theta <- # to write
## E step
res_Estep <- # to write
tau <- # to write
## check consistency
loglik[iter] <- # to write
Eloglik[iter] <- # to write
if (iter > 1)
cond <- (iter>=max.iter) | Eloglik[iter]-Eloglik[iter-1] < eps
}
res <- list(alpha = theta$alpha, mu = theta$mu, sigma = theta$sigma,
tau = tau, cl = apply(tau, 1, which.max),
Eloglik = Eloglik[1:iter],
loglik = loglik[1:iter])
res
}- Examples. We would like now to assess the performance of our EM algorithm. To do so, we generate:
mu1 <- 5 ; sigma1 <- 1; n1 <- 100
mu2 <- 10 ; sigma2 <- 1; n2 <- 200
mu3 <- 15 ; sigma3 <- 2; n3 <- 50
mu4 <- 20 ; sigma4 <- 3; n4 <- 100
cl <- rep(1:4,c(n1,n2,n3,n4))
x <- c(rnorm(n1,mu1,sigma1),rnorm(n2,mu2,sigma2),
rnorm(n3,mu3,sigma3),rnorm(n4,mu4,sigma4))
n <- length(x)
## we randomize the class ordering
rnd <- base::sample(1:n)
cl <- cl[rnd]
x <- x[rnd]
alpha <- c(n1,n2,n3,n4)/n
curve(alpha[1]*dnorm(x,mu1,sigma1) +
alpha[2]*dnorm(x,mu2,sigma2) +
alpha[3]*dnorm(x,mu3,sigma3) +
alpha[4]*dnorm(x,mu4,sigma3),
col="blue", lty=1, from=0,to=30, n=1000,
main="Theoretical Gaussian mixture and its components",
xlab="x", ylab="density")
curve(alpha[1]*dnorm(x,mu1,sigma1), col="red", add=TRUE, lty=2)
curve(alpha[2]*dnorm(x,mu2,sigma2), col="red", add=TRUE, lty=2)
curve(alpha[3]*dnorm(x,mu3,sigma3), col="red", add=TRUE, lty=2)
curve(alpha[4]*dnorm(x,mu4,sigma4), col="red", add=TRUE, lty=2)
rug(x)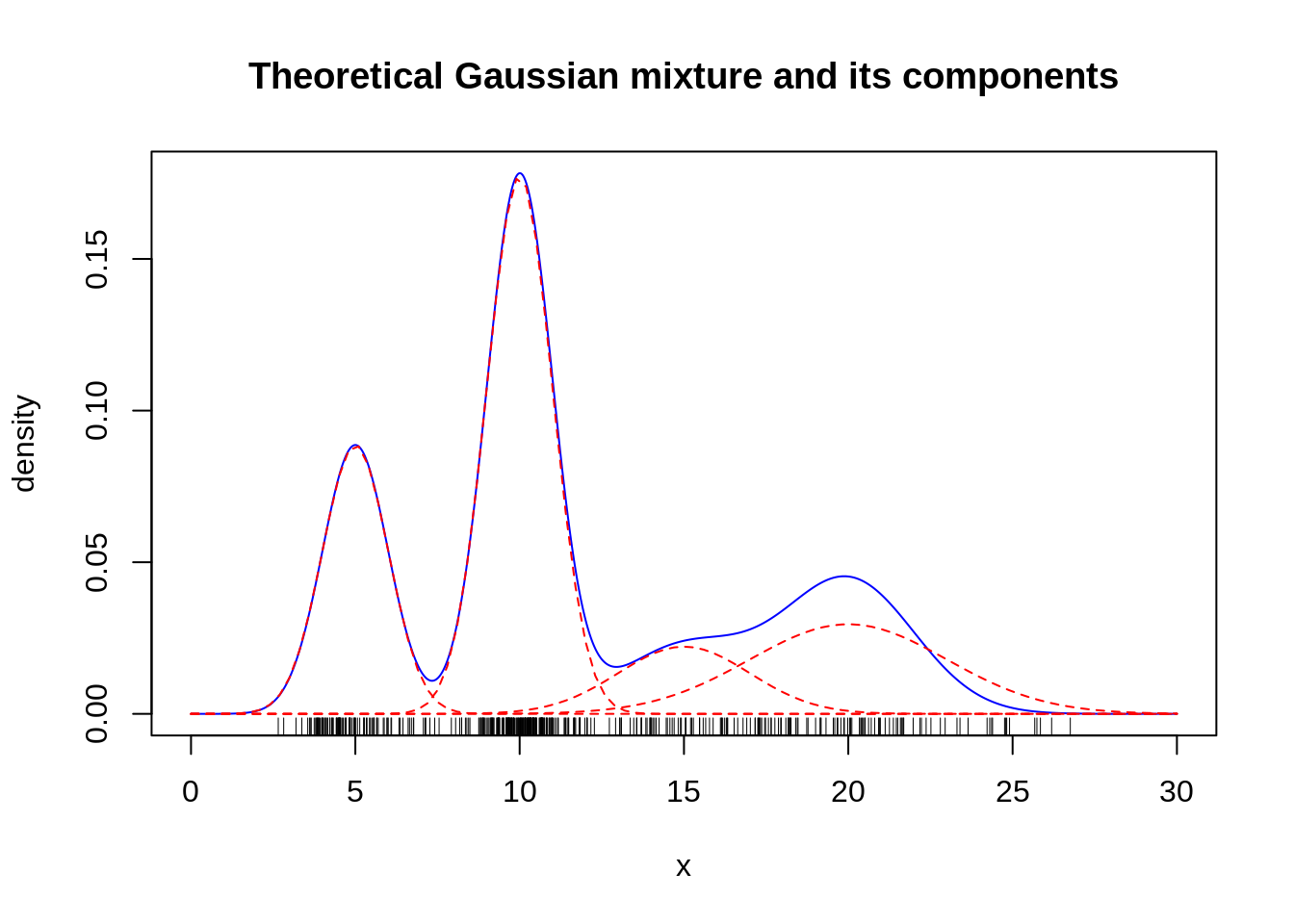 Try your EM algorithm on the simulated data. Comment the results. Consider different initialization. Compare with the output of the function normalmixEm in the package mixtools.
Try your EM algorithm on the simulated data. Comment the results. Consider different initialization. Compare with the output of the function normalmixEm in the package mixtools.
- Model Selection. Discuss the choice of \(Q\) by computing the BIC and ICL criterion and test it on your simulated data. \[ \mathrm{BIC}(Q) = - 2 \log L(X;\hat \alpha, \hat \mu, \hat \sigma^2) +\log(n) \mathrm{df}(Q). \] \[ \mathrm{ICL}(Q) = - 2 \log L(X,Z;\hat \alpha, \hat \mu, \hat \sigma^2) +\log(n) \mathrm{df}(Q). \]