Homework #3: correction
PCA for visualization, compression and exploratory analyses
MAP573 team
13/20/2020
Information
Data sets are available at https://github.com/jchiquet/MAP573/tree/master/data. Homework is due Sunday 10/18 23:59 in Rmd (see assignment in Moodle).
Package requirements
We start by loading a couple of packages for data manipulation, dimension reduction and fancy representations.
library(tidyverse) # advanced data manipulation and vizualisation
library(knitr) # R notebook export and formatting
library(FactoMineR) # Factor analysis
library(factoextra) # Fancy plotting of FactoMineR outputs
library(kableExtra) # integration of table in Rmarkdown
theme_set(theme_bw()) # set default ggplot2 theme to black and whiteSNP data: genotyping of various population
Data description
A single-nucleotide polymorphism is a substitution of a single nucleotide that occurs at a specific position in the genome, where each variation is present at a level of 0.5% from person to person in the population. They are coded as 0, 1 or 2 (meaning 0, 1 or 2 allels different regarding the reference population)
See the great wikipedia page for detail!
We can measure SNP for individuals with high trhoughput technology and SNP array. SNP chips for human contains more than 1 million variables! We only suggest to analyse a sample of a data set containing the 5500 most variant SNP for 728 individuals with various origin, with the following descriptors:
- CEU: Utah residents with Northern and Western European ancestry from the CEPH collection
- GIH: Gujarati Indians in Houston, Texas
- LWK: Luhya in Webuye, Kenya
- MKK: Maasai in Kinyawa, Kenya
- TSI: Toscani in Italia
- YRI: Yoruba in Ibadan, Nigeria
The data are imported as follows:
load("data/SNP.RData")
snp <- data$Geno %>% as_tibble() %>%
add_column(origin = data$origin, .before = 1) Questions
- Have a look at the data a make some brief descriptive plot/summary
- Fit a PCA on these data. Justify the scaling or not.
- Represent the scree plot for the 100 first axes. Comment. Estimate the number of axis with
estim_npc - Plot individual factor maps on axes 1, 2 and 3. Add colors and ellipses associated with the factor ‘origin’
- Check who are the first, say, 250 most contributive individuals to these axes. Show them in the projection. Have a look at the cosine of the most influent guys.
- Plot the correlation circle. Only retain variables based on the quality of their representation and/or degree of contribution to the axes represented.
- Summarize the above analyses in biplots. Add fancy colors and whatever.
- Indicate a group of individual as supplementary (i.e., not use to fit the PC). Show how excluding a group influence (or not) the fit and the projection, by exploring several groups. Explain.
Solution
- Have a look at the data a make some brief descriptive plot/summary
The first column is a categorical variable describing the orgin of each individual, with details on the acronyme given above
barplot(table(snp$origin), las = 3, main = 'distribution of origin')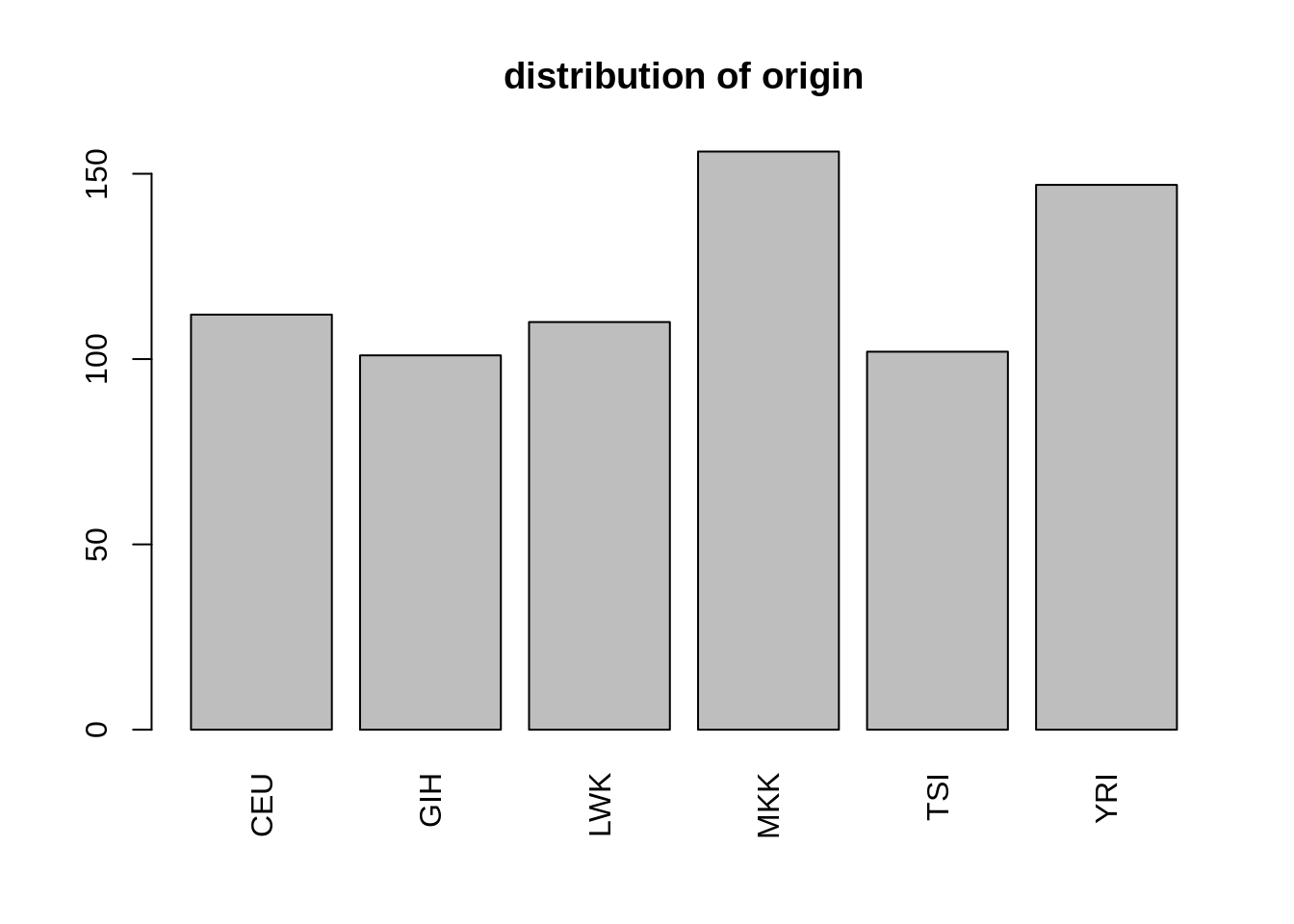
- Fit a PCA on these data. Justify the scaling or not.
I do not scale, since SNP value are suppose to live on the same scale (values in \(\{0, 1, 2\}\)).
acp <- PCA(snp, quali.sup = 1, scale.unit = FALSE, graph = FALSE, ncp = 500)## Warning in PCA(snp, quali.sup = 1, scale.unit = FALSE, graph = FALSE, ncp =
## 500): Missing values are imputed by the mean of the variable: you should use the
## imputePCA function of the missMDA package- Represent the scree plot for the 100 first axes. Comment
First axes are more informative than the other, and then the information is generally well spread.
fviz_eig(acp, ncp = 100)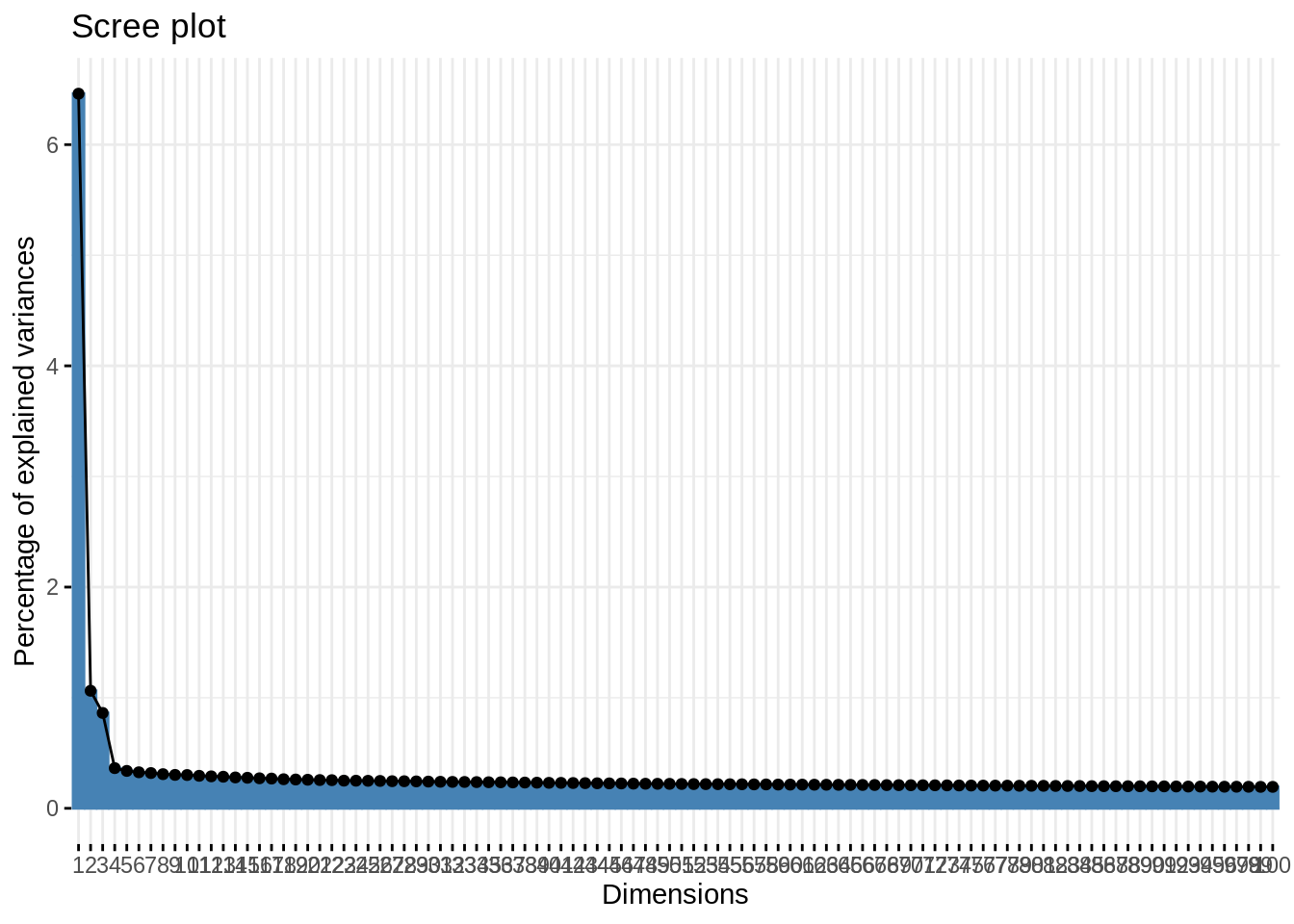
npc <- select(snp, -origin) %>% replace(is.na(.), 0) %>%
as.matrix() %>% scale(TRUE, FALSE) %>%
estim_ncp(ncp.max = 15, scale = FALSE)
plot(npc$criterion, type = "b", xlab = 'number of component', ylab = 'GCV')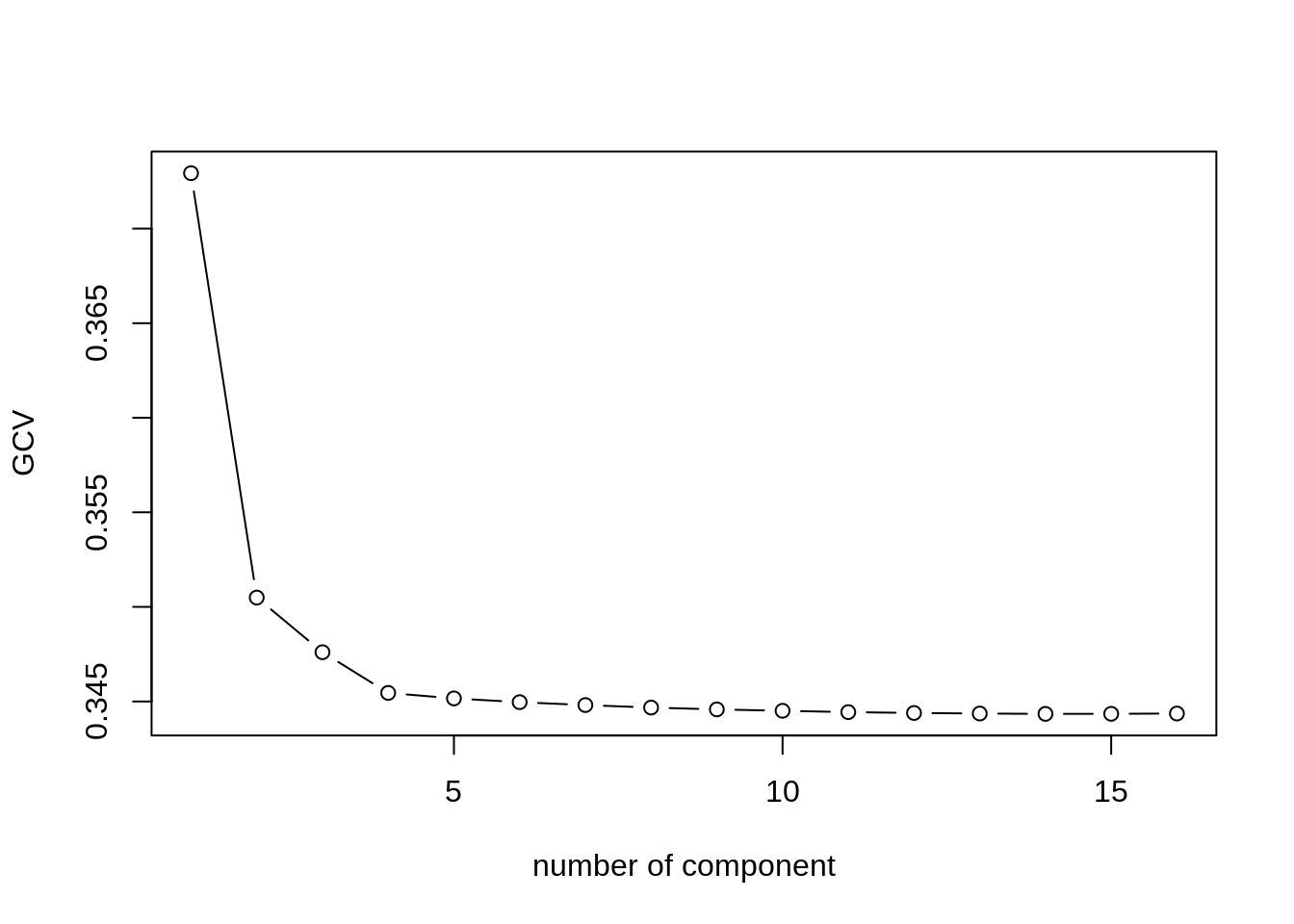
- Plot individual factor maps on axes 1, 2 and 3. Add color and ellipse associated with the origin
Argument habillage or col.ind will have the same effect, but the first will be more useful later.
Impressive how the population are well separated!
fviz_pca_ind(acp, habillage = "origin", addEllipses = TRUE)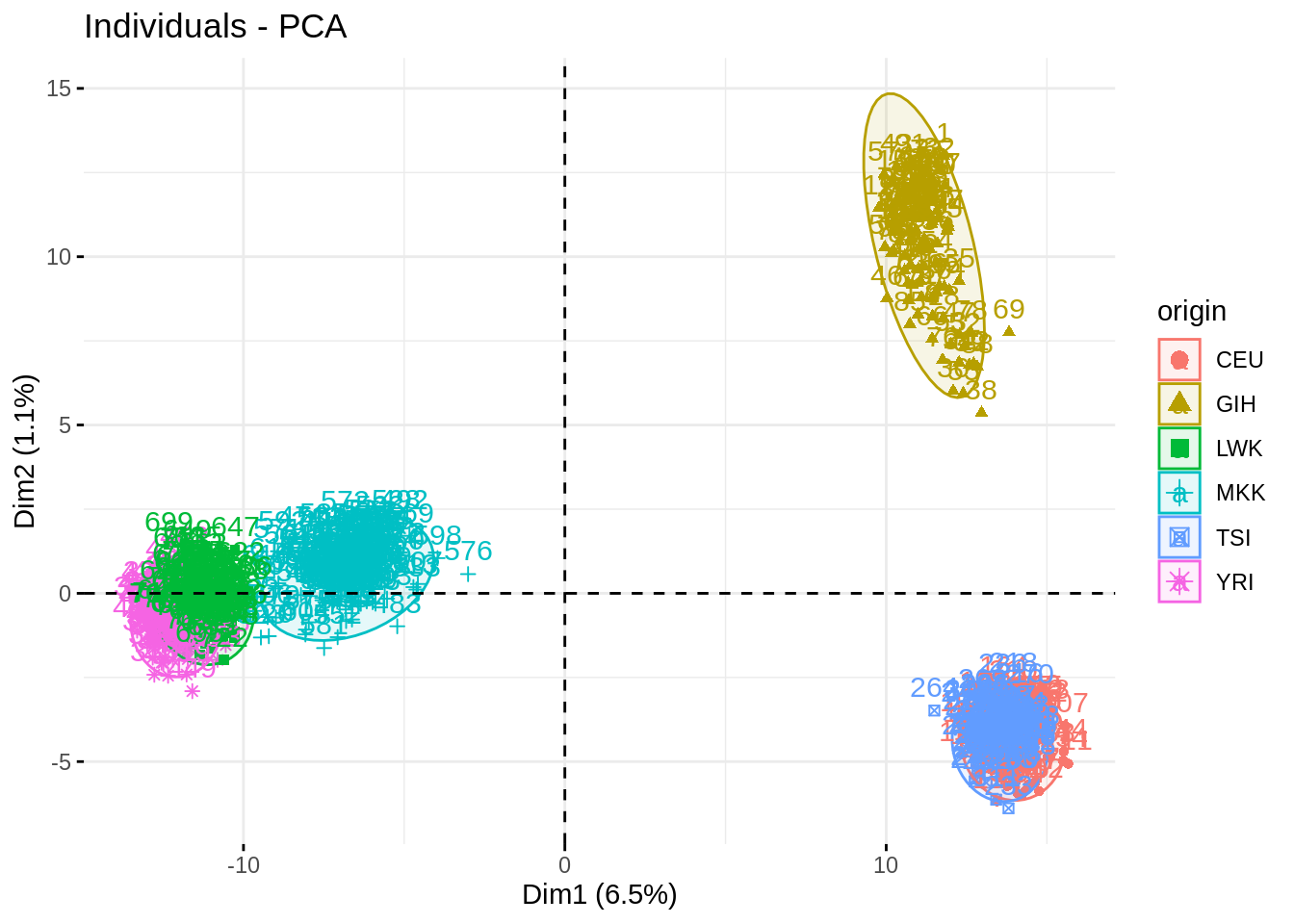
fviz_pca_ind(acp, habillage = "origin", axes = c(1,3), addEllipses = TRUE)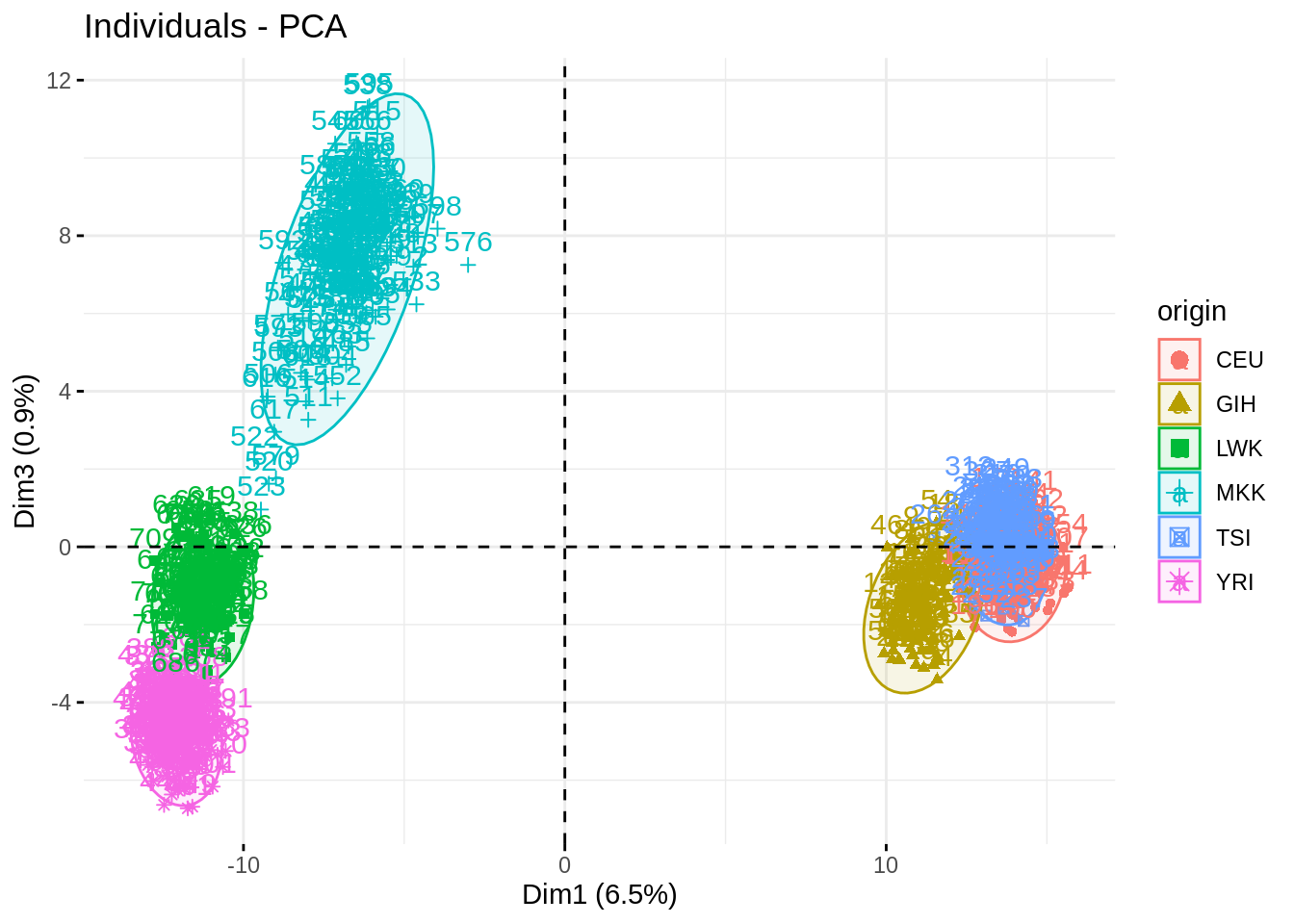
fviz_pca_ind(acp, habillage = "origin", axes = c(2,3), addEllipses = TRUE)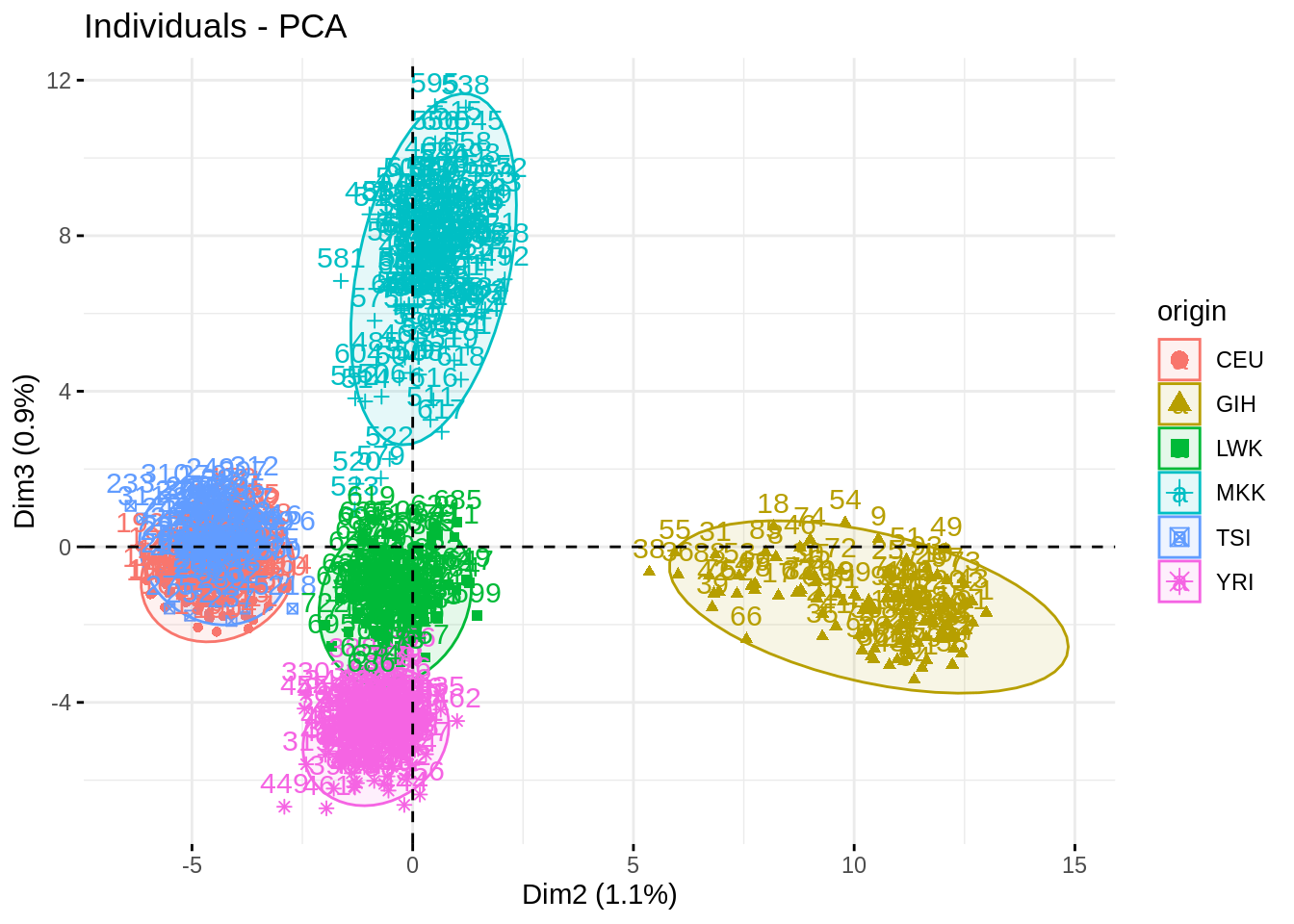
- Check who are the first, say, 250 most contributive individual to these axes. Show them in the projection. Have a look at the cosine of the most influent guys.
fviz_contrib(acp, "ind", axes = 1:3, top = 250)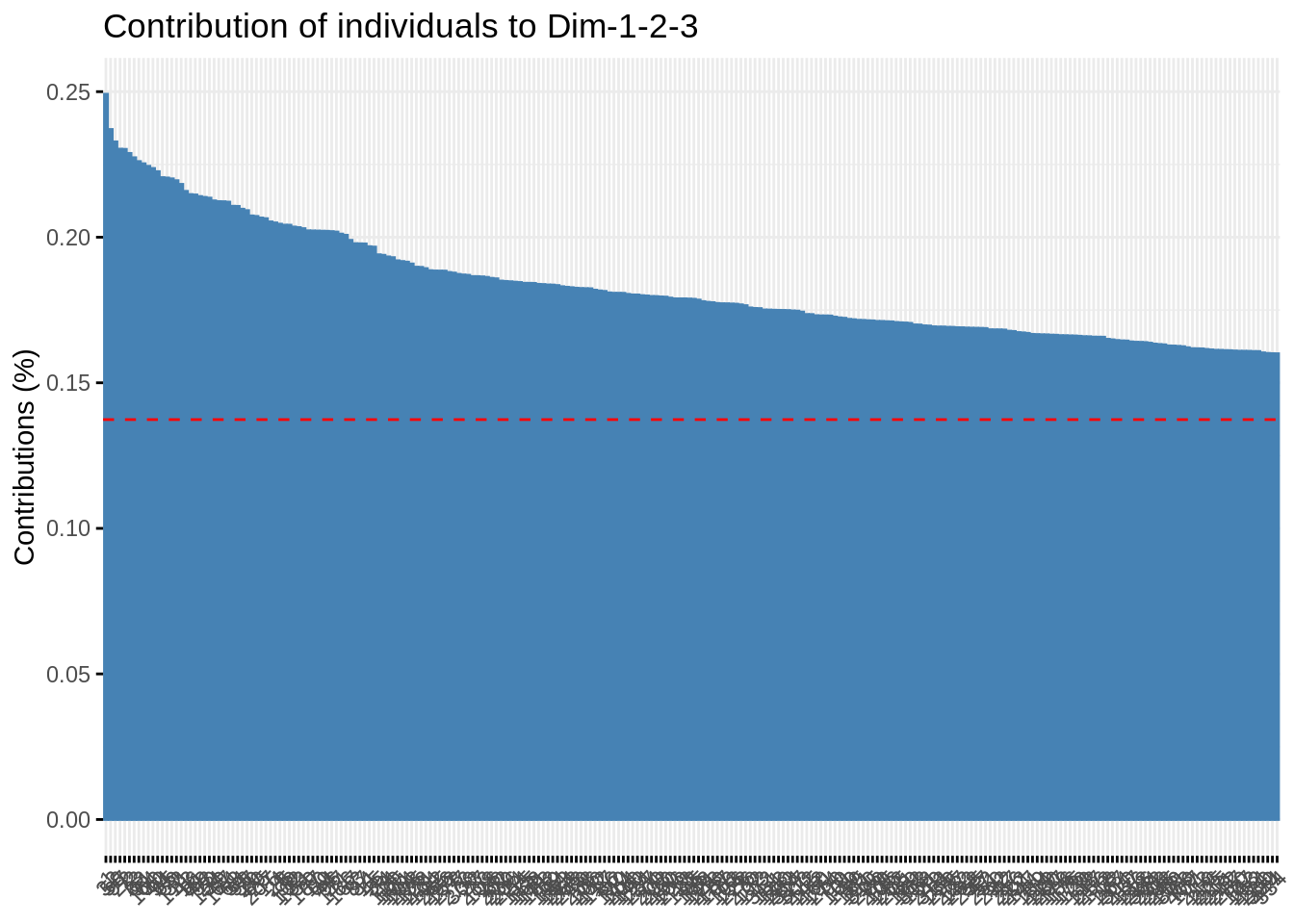
fviz_pca_ind(acp, habillage = "origin", select.ind = list(contrib = 250))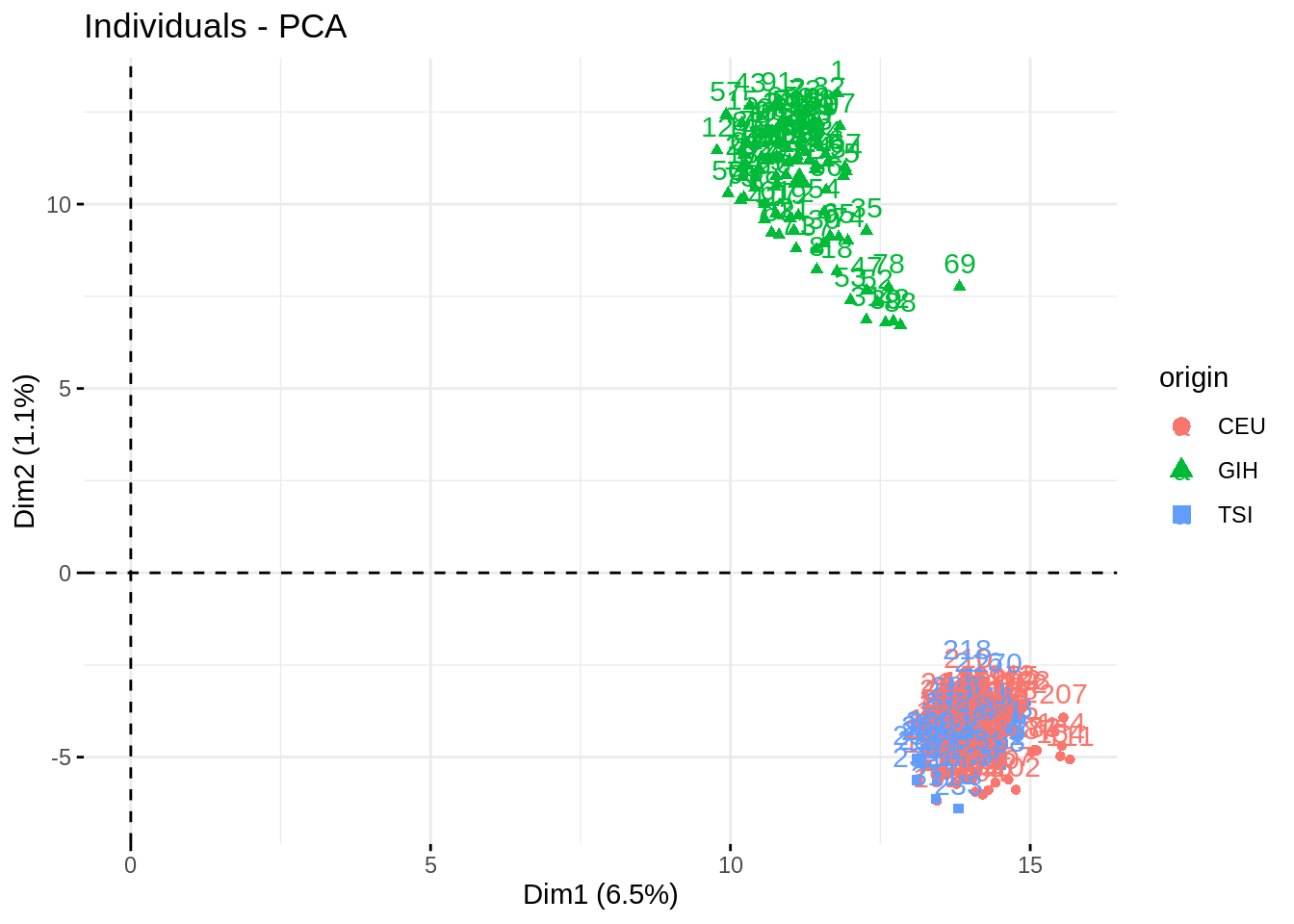
fviz_pca_ind(acp, axes = c(2,3), habillage = "origin", select.ind = list(contrib = 250))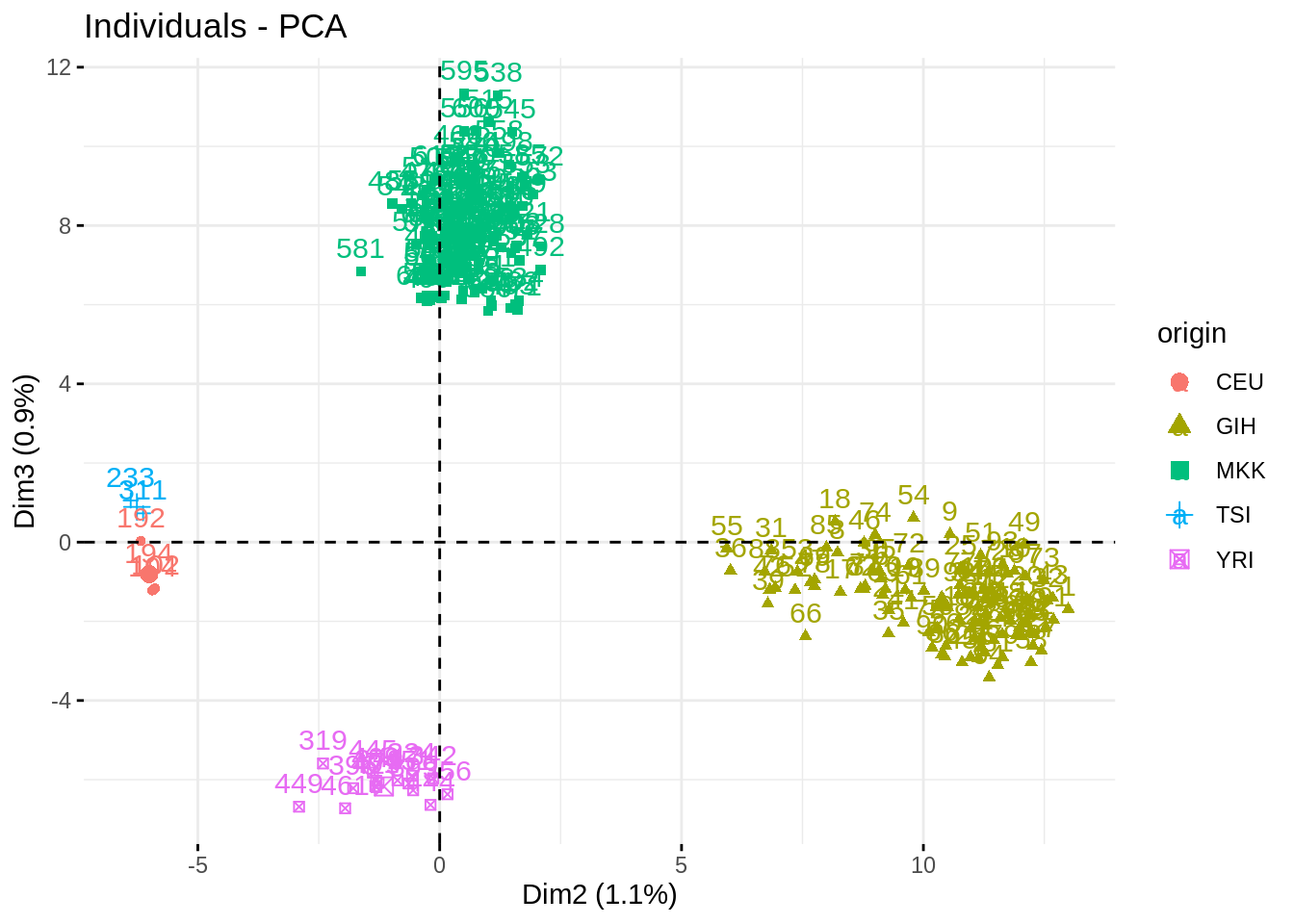
fviz_pca_ind(acp, habillage = "origin", select.ind = list(contrib = 250))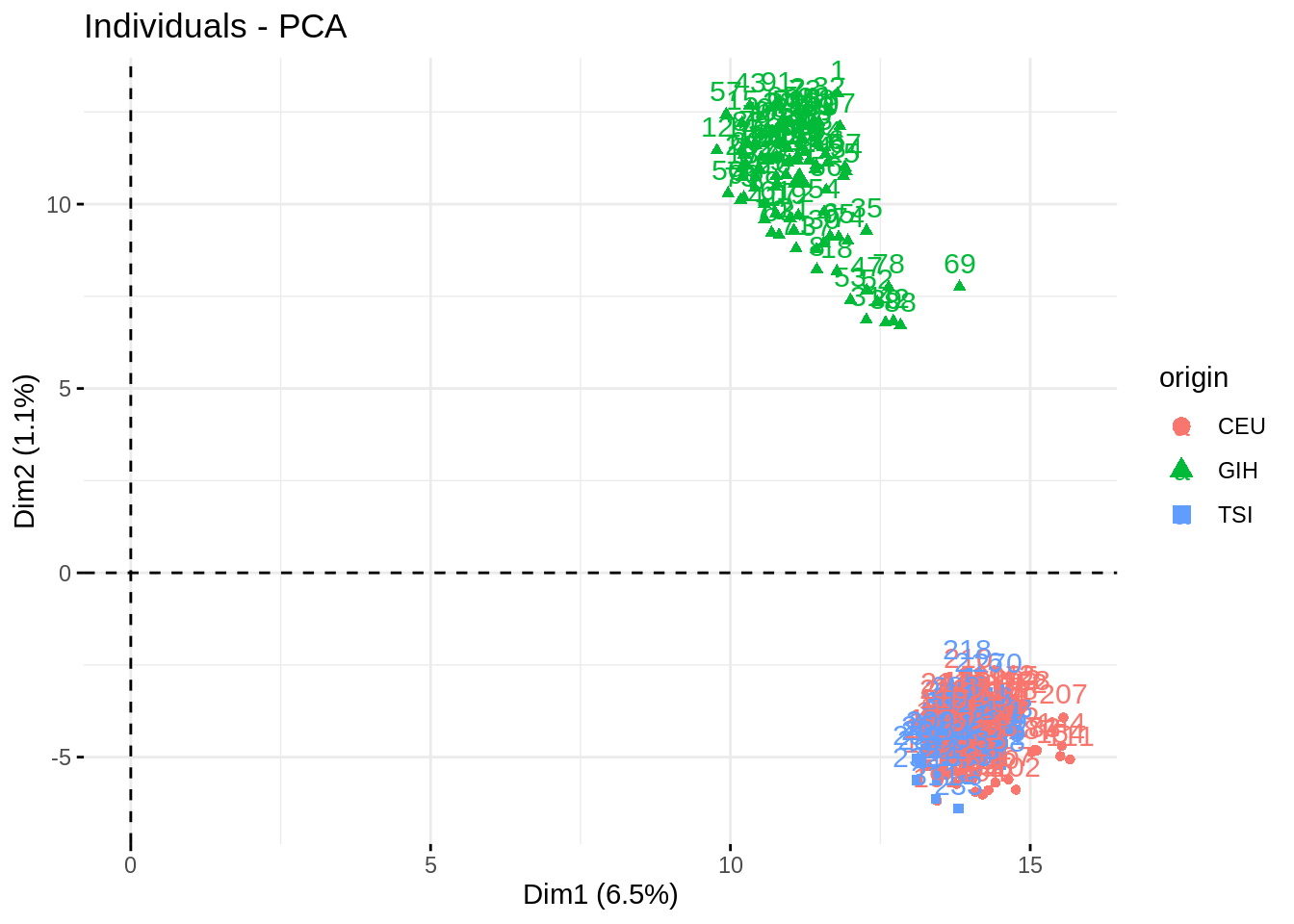
fviz_pca_ind(acp, habillage = "origin", select.ind = list(contrib = 250))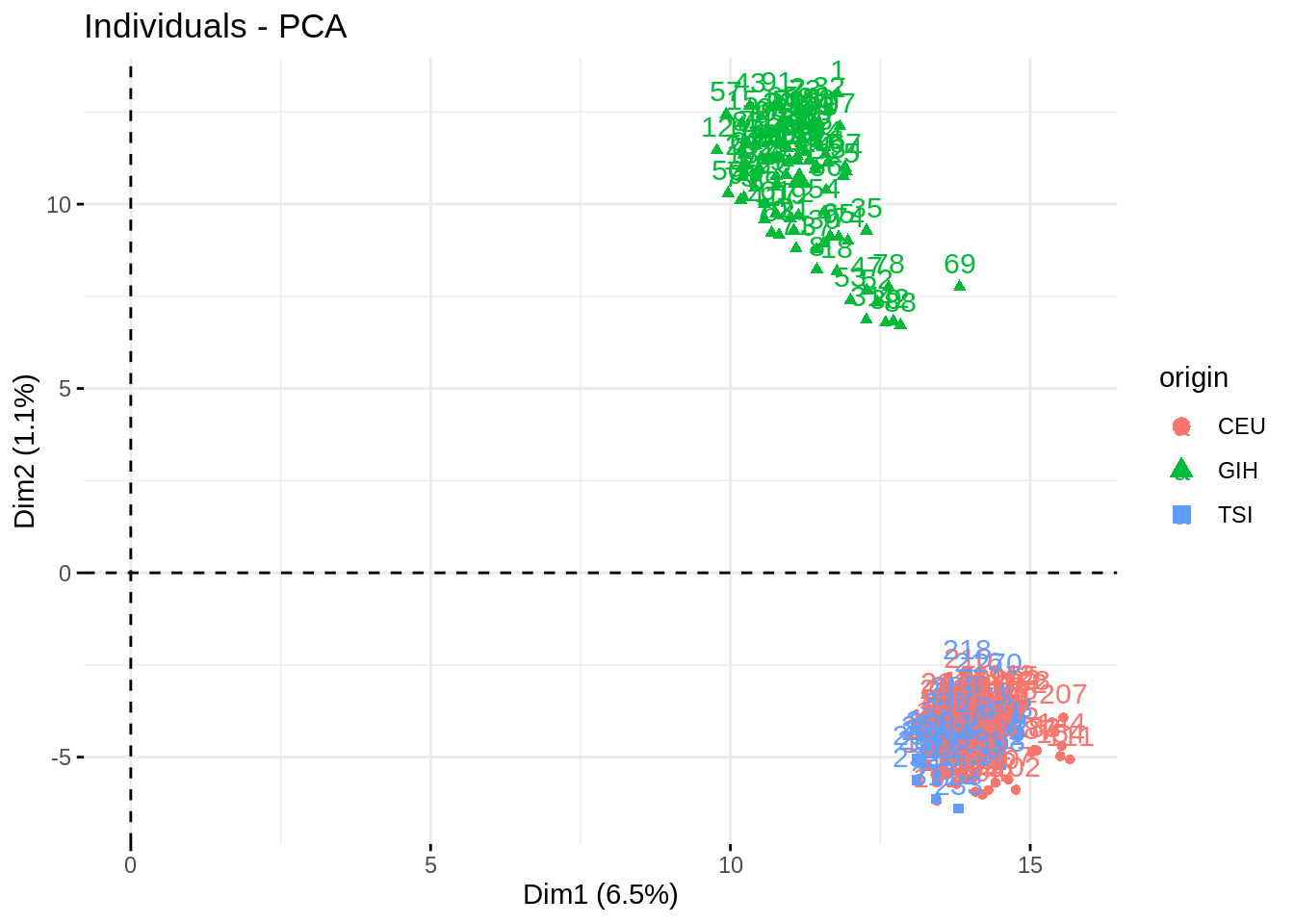
- Plot the correlation circle. Only retain variables based on the quality of their representation and/or degree of contribution to the axes represented.
fviz_contrib(acp, "var", axes = 1:3, top = 500)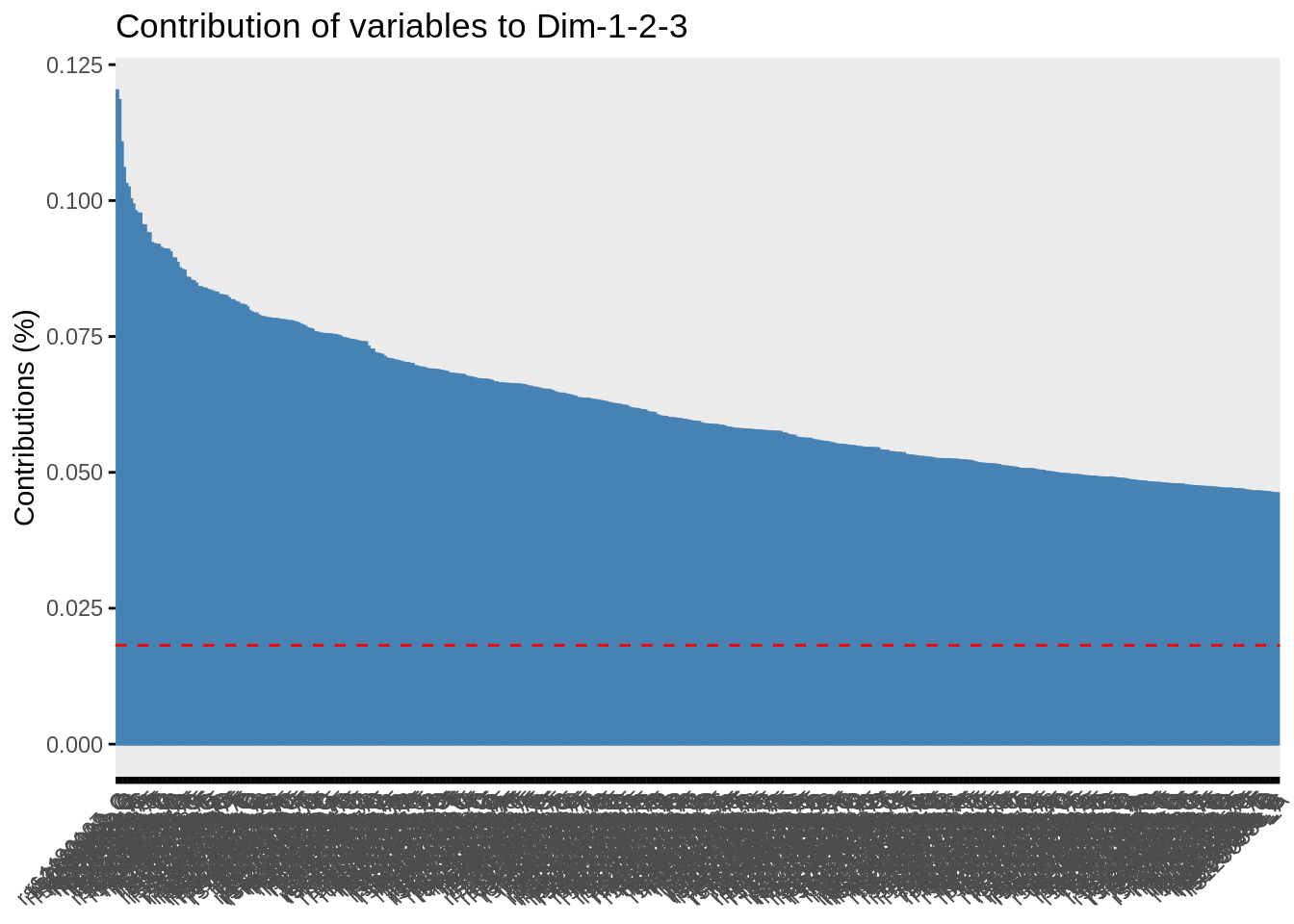
fviz_pca_var(acp, select.var = list(contrib = 50), col.var = 'cos2')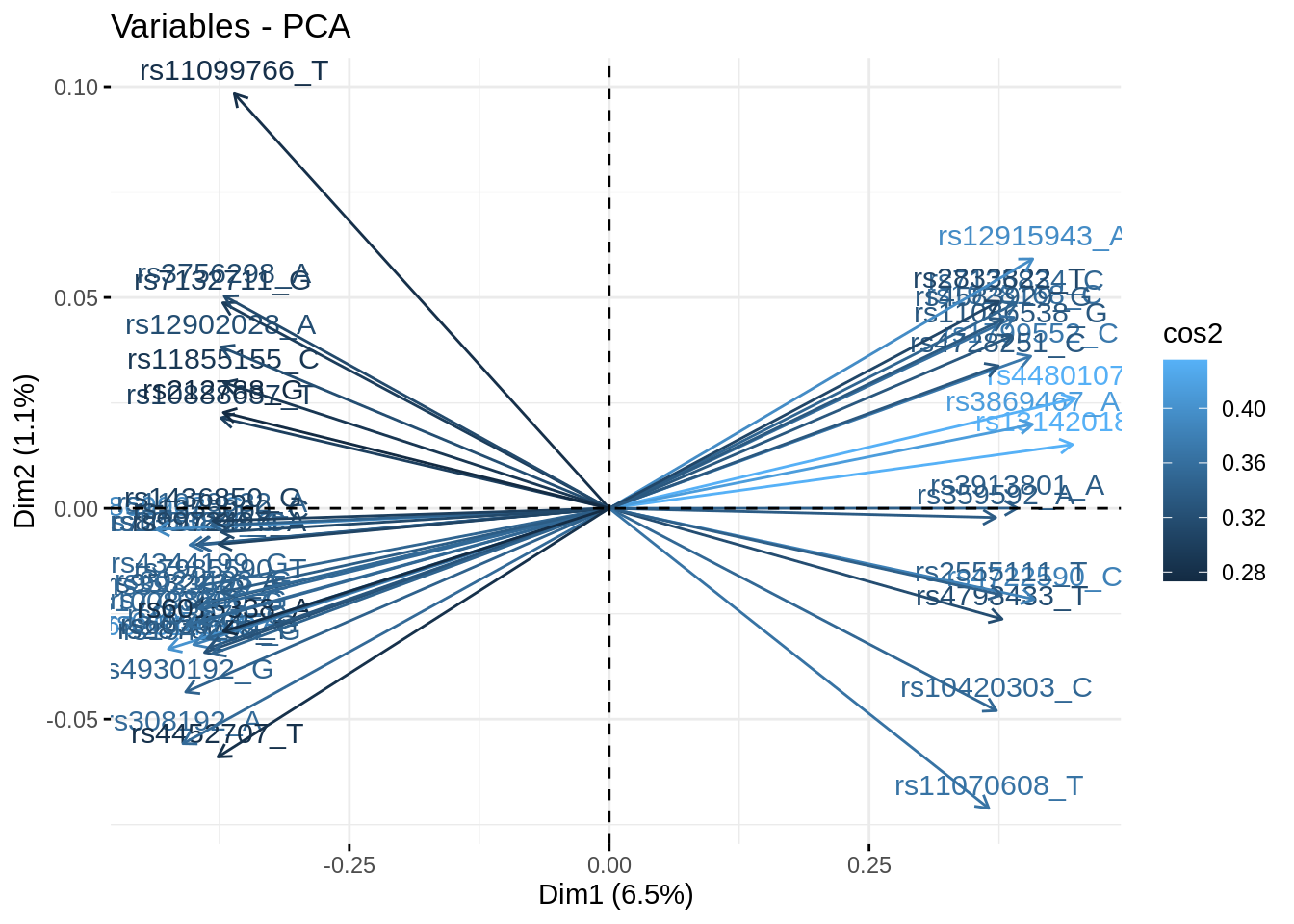
- Summarize the above analyses in biplots. Add fancy colors
Just example, you can do better/different than that!
fviz_pca_biplot(acp, axes = c(1,3), habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 40))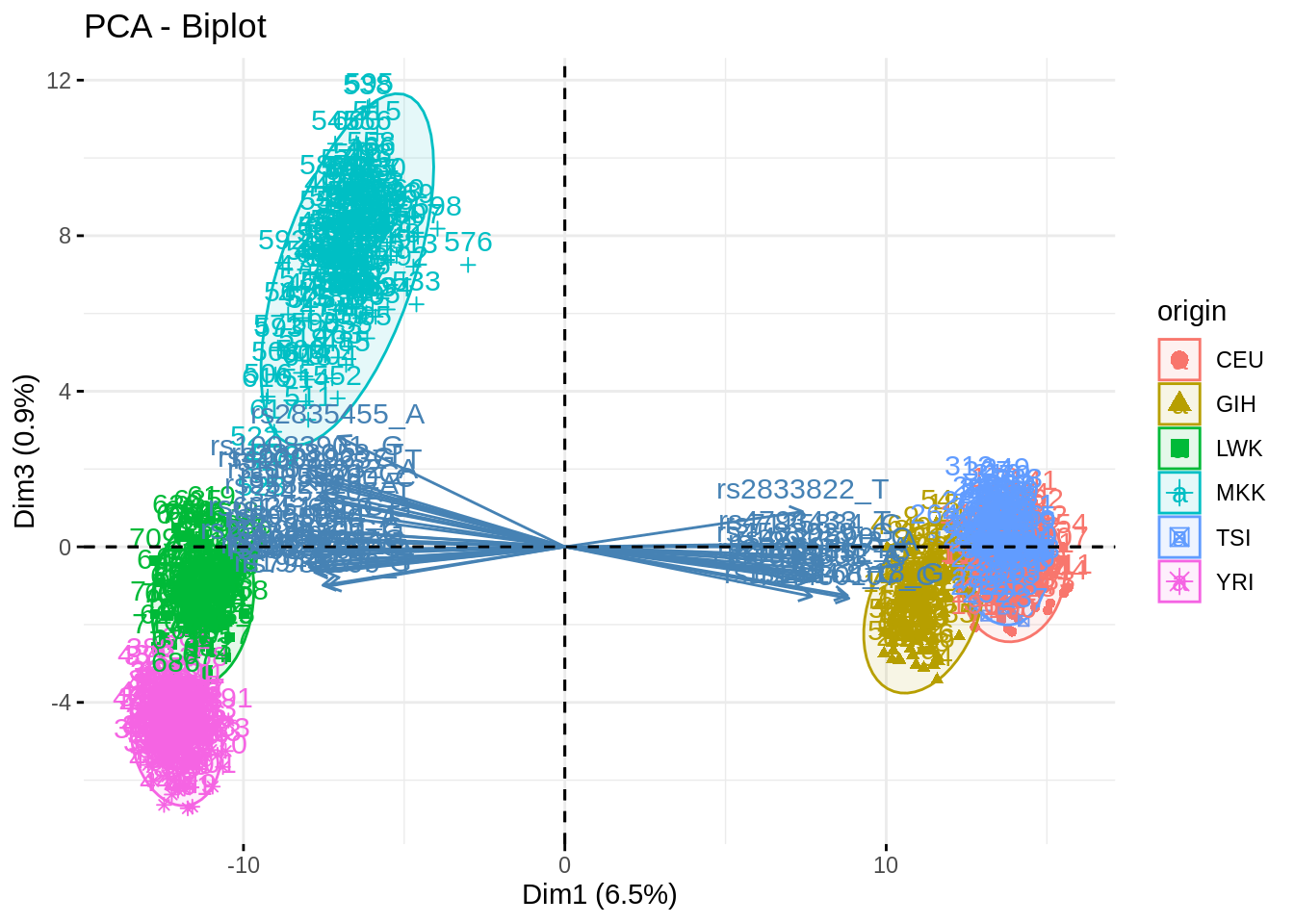
fviz_pca_biplot(acp, axes = c(1,3), habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 40))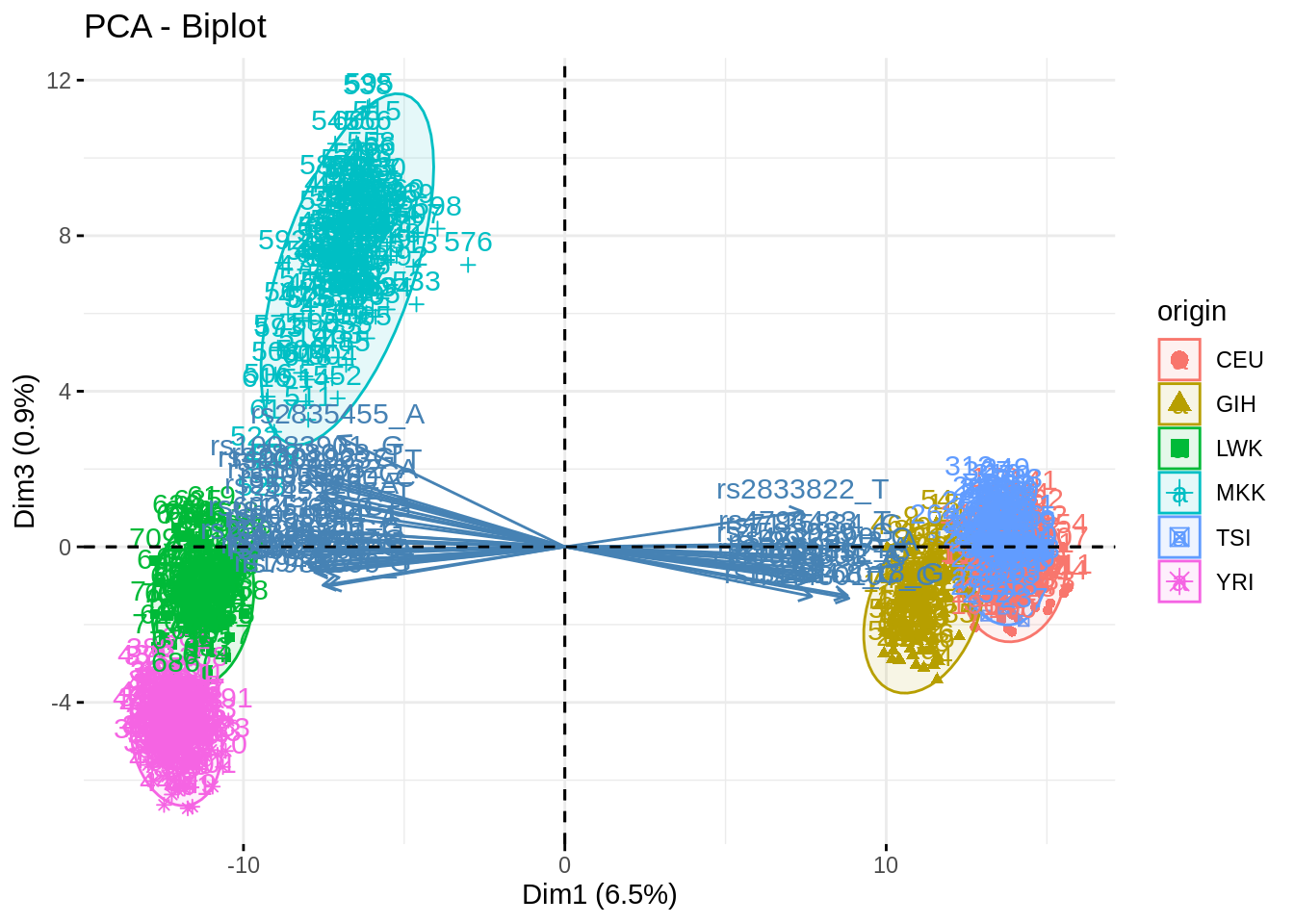
fviz_pca_biplot(acp, axes = c(2,3), habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 40))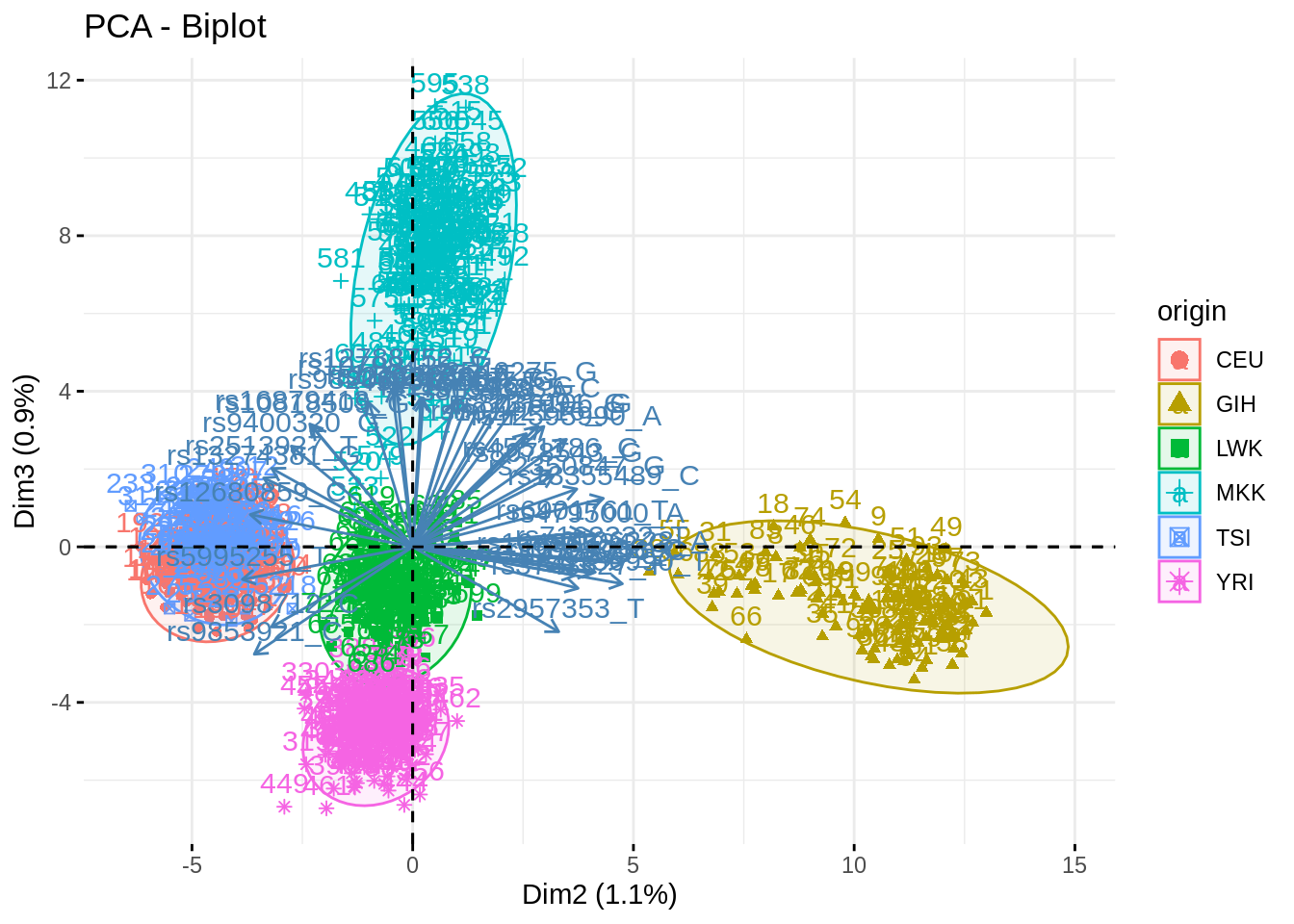
- Indicate a group of individual as supplmentary (i.e., not use to fit the PC). Show how excluding a group influence (or not) the fit and the projection, by exploring several groups. Explain
Depending on the proximity of the group to the cloud and to some particular existing groups, the fit is more or less altered.
acp_noMKK <- PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "MKK"), scale.unit = FALSE, graph = FALSE, ncp = 500)## Warning in PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "MKK"), :
## Missing values are imputed by the mean of the variable: you should use the
## imputePCA function of the missMDA packagefviz_pca_biplot(acp, habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 40), col.ind.sup = "black")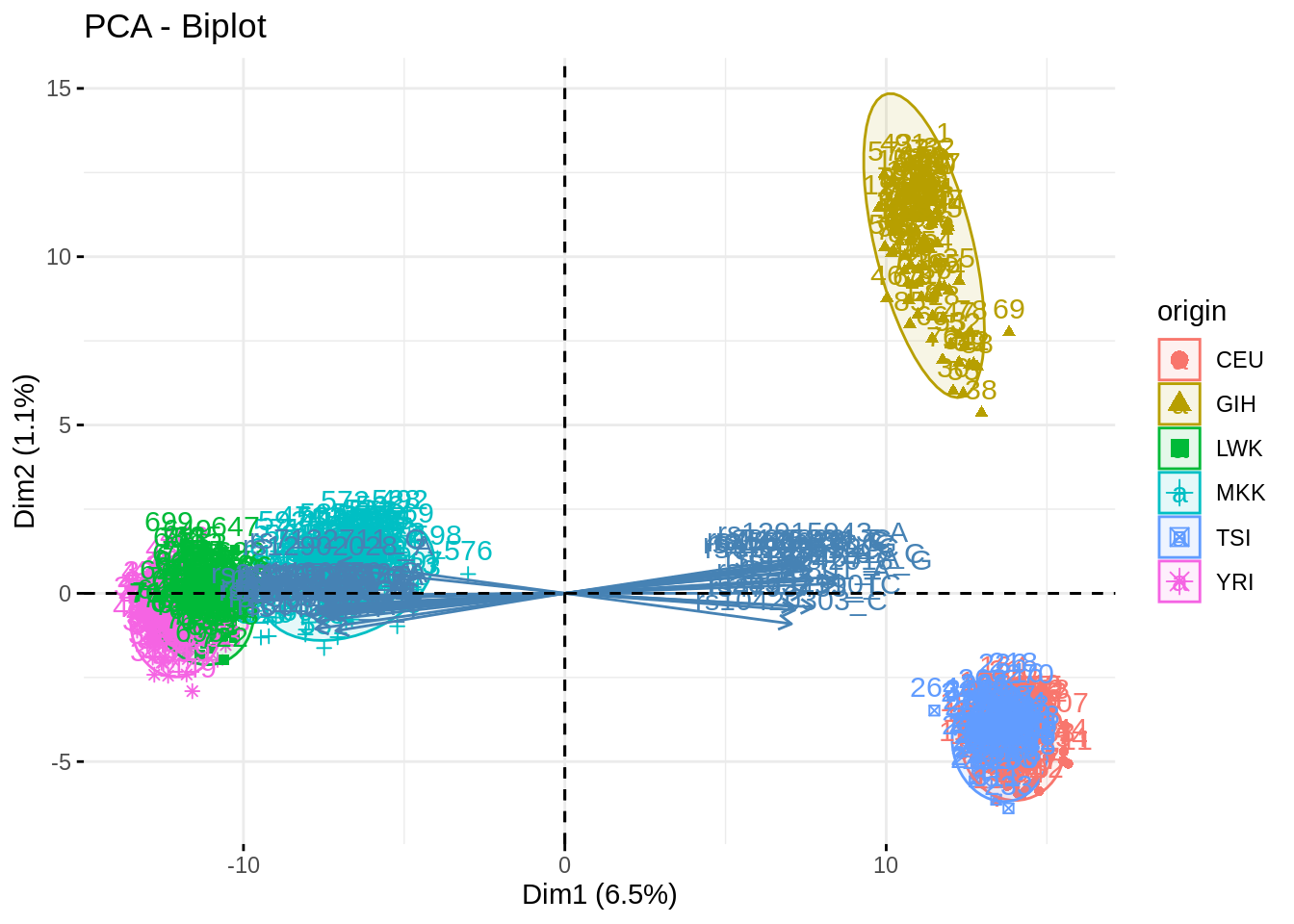
acp_noTSI <- PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "TSI"), scale.unit = FALSE, graph = FALSE, ncp = 500)## Warning in PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "TSI"), :
## Missing values are imputed by the mean of the variable: you should use the
## imputePCA function of the missMDA packagefviz_pca_biplot(acp, habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 40), col.ind.sup = "black")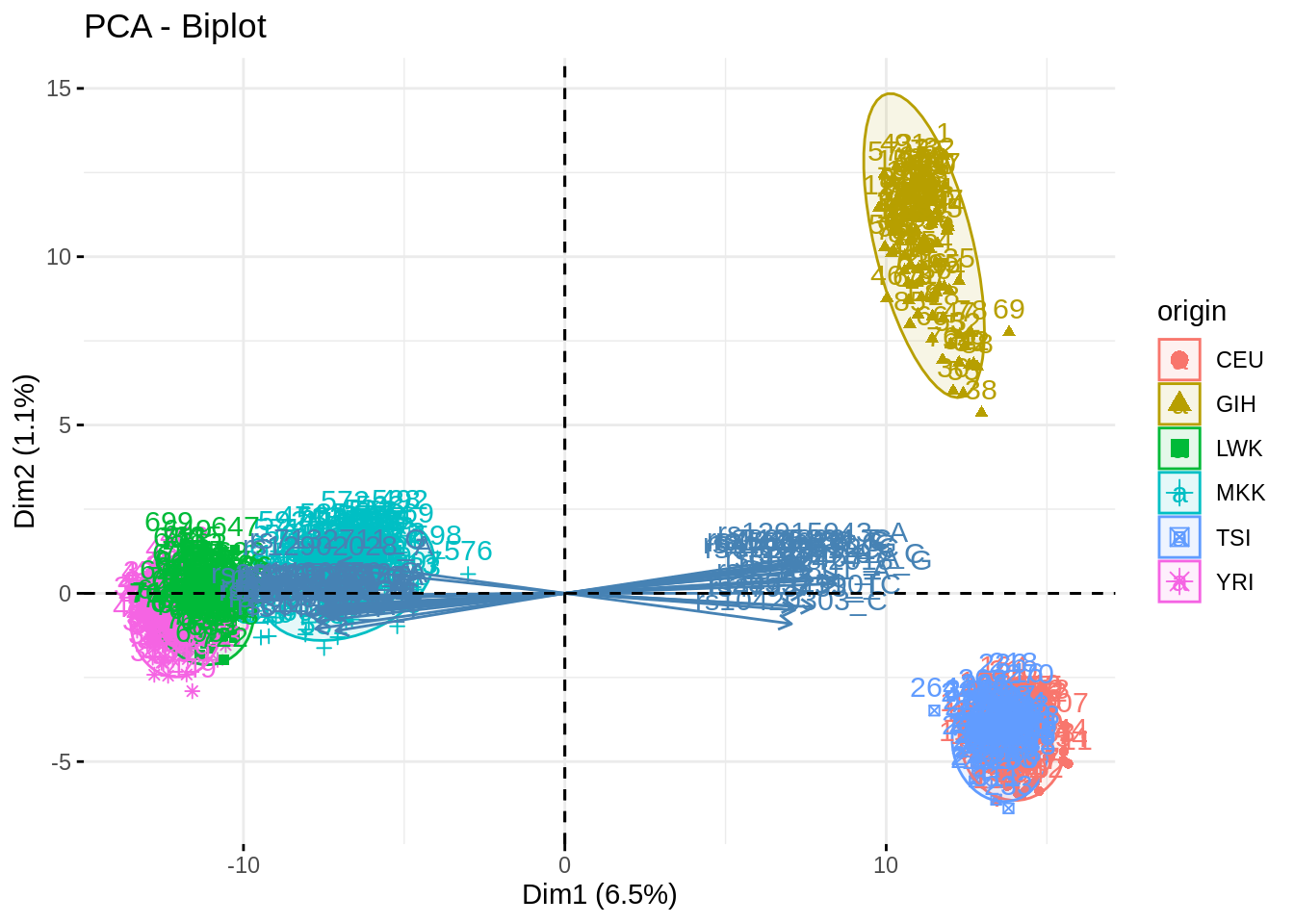
acp_noGIH <- PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "GIH"), scale.unit = FALSE, graph = FALSE, ncp = 500)## Warning in PCA(snp, quali.sup = 1, ind.sup = which(snp$origin == "GIH"), :
## Missing values are imputed by the mean of the variable: you should use the
## imputePCA function of the missMDA packagefviz_pca_biplot(acp, habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 25), col.ind.sup = "black")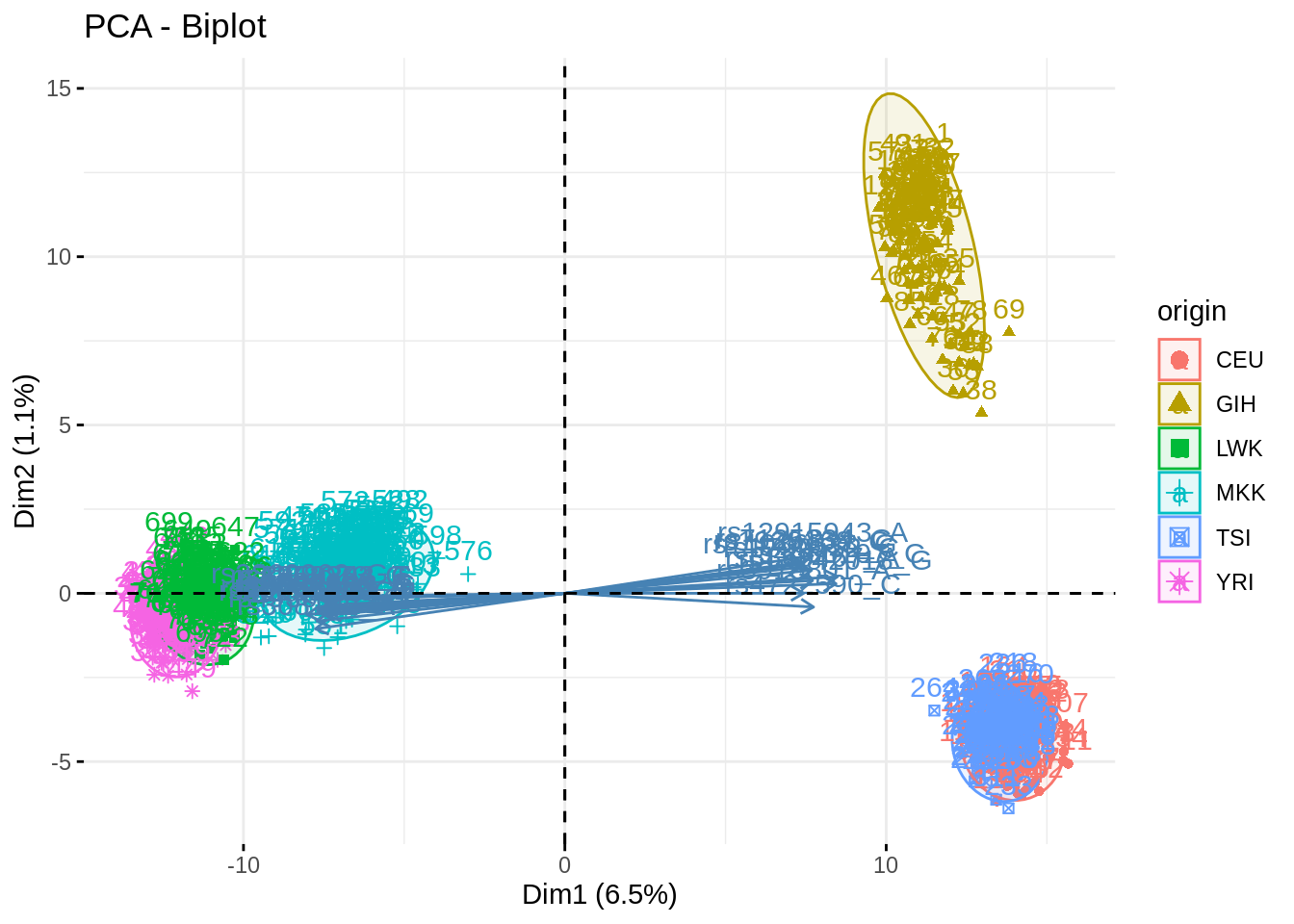
fviz_pca_biplot(acp, axes = c(2,3), habillage = "origin", addEllipses = TRUE, select.var = list(contrib = 25), col.ind.sup = "black")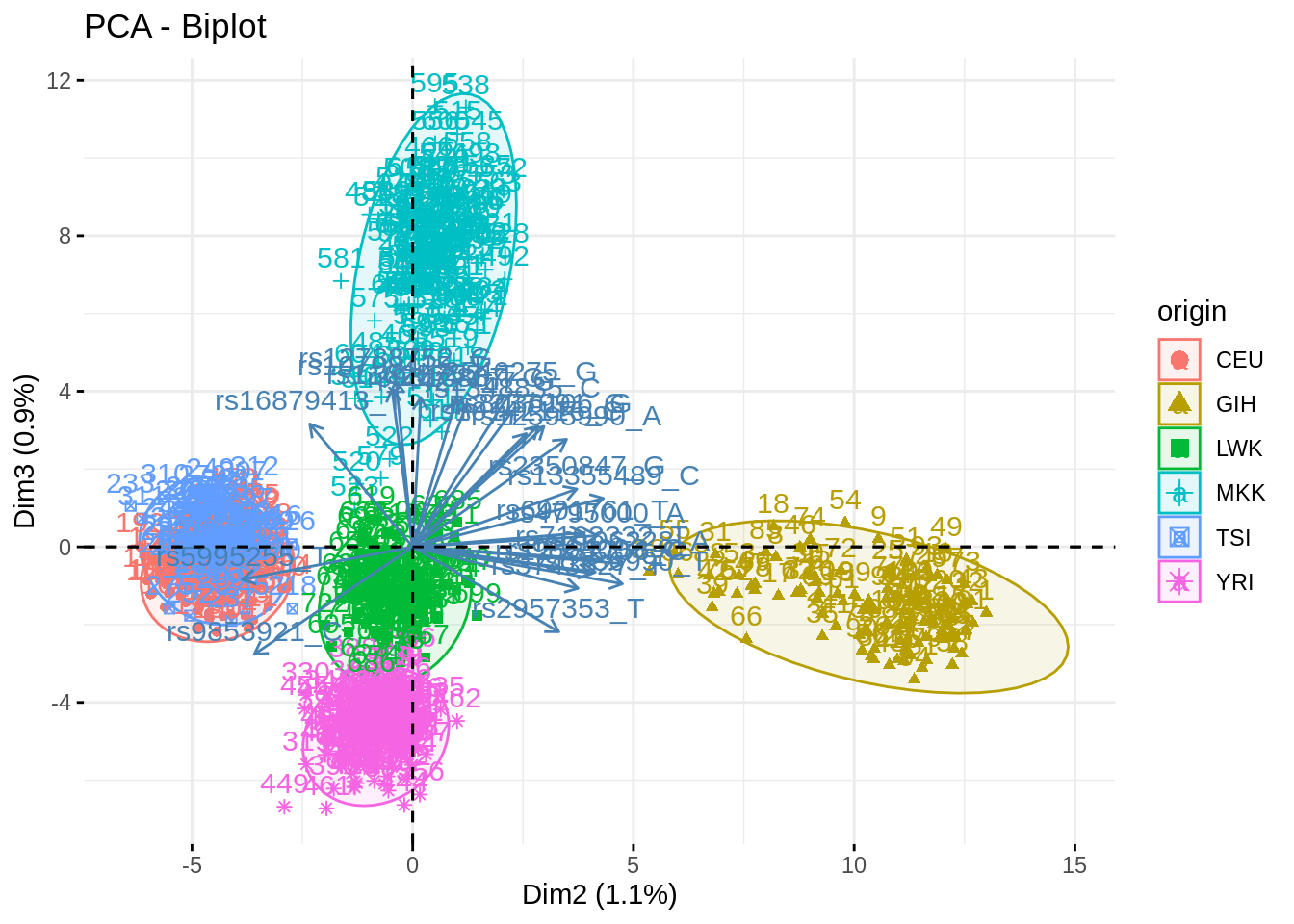
MNIST data: handwritten digit data
Data description
The MNIST dataset is an acronym that stands for the Modified National Institute of Standards and Technology dataset. It is a dataset of 60,000 small square 28×28 pixel grayscale images of handwritten single digits between 0 and 9. It is commonly used for training various image processing systems. The database is also widely used for training and testing in the field of machine learning.
The full data set is available here.
But careful!!, it weigths more than 100 Mo so check your internet connection
A sample of 2,000 instances is provided for convenience mnist_sample.
Questions
- Load an format the data (first column is the label) as a
tibbleor adata.frame. Each row is a vector with size 784 (28 x 28) + a column for the label. - Write a function to plot an 28 x 28 image based on a vector, and test it on a row of your data frame.
- Make a PCA, choice whether to scale or not.
- Make the usual diagnostic plots (screeplot, individual, variable) for the first axes, and comment. Use GCV to select the number of axes.
- Write a function to reconstruct an image with two argument: \(i\) (an instance) \(k\) (the number of component used).
- Plot the reconstructed image for various value of \(k\)
Solution
- Load an format the data (first column is the label) as a
tibbleor adata.frame. Each row is a vector with size 784 (28 x 28) + a column for the label.
mnist_raw <- read_csv("data/mnist_sample.csv", col_names = FALSE, n_max = 2000)##
## ── Column specification ────────────────────────────────────────────────────────
## cols(
## .default = col_double()
## )
## ℹ Use `spec()` for the full column specifications.mnist <- mnist_raw %>%
rename(label = X1) %>%
mutate(instance = row_number())
mnist %>% head() %>% kable() %>% kable_styling() %>%
scroll_box(width = "100%")| label | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 | X13 | X14 | X15 | X16 | X17 | X18 | X19 | X20 | X21 | X22 | X23 | X24 | X25 | X26 | X27 | X28 | X29 | X30 | X31 | X32 | X33 | X34 | X35 | X36 | X37 | X38 | X39 | X40 | X41 | X42 | X43 | X44 | X45 | X46 | X47 | X48 | X49 | X50 | X51 | X52 | X53 | X54 | X55 | X56 | X57 | X58 | X59 | X60 | X61 | X62 | X63 | X64 | X65 | X66 | X67 | X68 | X69 | X70 | X71 | X72 | X73 | X74 | X75 | X76 | X77 | X78 | X79 | X80 | X81 | X82 | X83 | X84 | X85 | X86 | X87 | X88 | X89 | X90 | X91 | X92 | X93 | X94 | X95 | X96 | X97 | X98 | X99 | X100 | X101 | X102 | X103 | X104 | X105 | X106 | X107 | X108 | X109 | X110 | X111 | X112 | X113 | X114 | X115 | X116 | X117 | X118 | X119 | X120 | X121 | X122 | X123 | X124 | X125 | X126 | X127 | X128 | X129 | X130 | X131 | X132 | X133 | X134 | X135 | X136 | X137 | X138 | X139 | X140 | X141 | X142 | X143 | X144 | X145 | X146 | X147 | X148 | X149 | X150 | X151 | X152 | X153 | X154 | X155 | X156 | X157 | X158 | X159 | X160 | X161 | X162 | X163 | X164 | X165 | X166 | X167 | X168 | X169 | X170 | X171 | X172 | X173 | X174 | X175 | X176 | X177 | X178 | X179 | X180 | X181 | X182 | X183 | X184 | X185 | X186 | X187 | X188 | X189 | X190 | X191 | X192 | X193 | X194 | X195 | X196 | X197 | X198 | X199 | X200 | X201 | X202 | X203 | X204 | X205 | X206 | X207 | X208 | X209 | X210 | X211 | X212 | X213 | X214 | X215 | X216 | X217 | X218 | X219 | X220 | X221 | X222 | X223 | X224 | X225 | X226 | X227 | X228 | X229 | X230 | X231 | X232 | X233 | X234 | X235 | X236 | X237 | X238 | X239 | X240 | X241 | X242 | X243 | X244 | X245 | X246 | X247 | X248 | X249 | X250 | X251 | X252 | X253 | X254 | X255 | X256 | X257 | X258 | X259 | X260 | X261 | X262 | X263 | X264 | X265 | X266 | X267 | X268 | X269 | X270 | X271 | X272 | X273 | X274 | X275 | X276 | X277 | X278 | X279 | X280 | X281 | X282 | X283 | X284 | X285 | X286 | X287 | X288 | X289 | X290 | X291 | X292 | X293 | X294 | X295 | X296 | X297 | X298 | X299 | X300 | X301 | X302 | X303 | X304 | X305 | X306 | X307 | X308 | X309 | X310 | X311 | X312 | X313 | X314 | X315 | X316 | X317 | X318 | X319 | X320 | X321 | X322 | X323 | X324 | X325 | X326 | X327 | X328 | X329 | X330 | X331 | X332 | X333 | X334 | X335 | X336 | X337 | X338 | X339 | X340 | X341 | X342 | X343 | X344 | X345 | X346 | X347 | X348 | X349 | X350 | X351 | X352 | X353 | X354 | X355 | X356 | X357 | X358 | X359 | X360 | X361 | X362 | X363 | X364 | X365 | X366 | X367 | X368 | X369 | X370 | X371 | X372 | X373 | X374 | X375 | X376 | X377 | X378 | X379 | X380 | X381 | X382 | X383 | X384 | X385 | X386 | X387 | X388 | X389 | X390 | X391 | X392 | X393 | X394 | X395 | X396 | X397 | X398 | X399 | X400 | X401 | X402 | X403 | X404 | X405 | X406 | X407 | X408 | X409 | X410 | X411 | X412 | X413 | X414 | X415 | X416 | X417 | X418 | X419 | X420 | X421 | X422 | X423 | X424 | X425 | X426 | X427 | X428 | X429 | X430 | X431 | X432 | X433 | X434 | X435 | X436 | X437 | X438 | X439 | X440 | X441 | X442 | X443 | X444 | X445 | X446 | X447 | X448 | X449 | X450 | X451 | X452 | X453 | X454 | X455 | X456 | X457 | X458 | X459 | X460 | X461 | X462 | X463 | X464 | X465 | X466 | X467 | X468 | X469 | X470 | X471 | X472 | X473 | X474 | X475 | X476 | X477 | X478 | X479 | X480 | X481 | X482 | X483 | X484 | X485 | X486 | X487 | X488 | X489 | X490 | X491 | X492 | X493 | X494 | X495 | X496 | X497 | X498 | X499 | X500 | X501 | X502 | X503 | X504 | X505 | X506 | X507 | X508 | X509 | X510 | X511 | X512 | X513 | X514 | X515 | X516 | X517 | X518 | X519 | X520 | X521 | X522 | X523 | X524 | X525 | X526 | X527 | X528 | X529 | X530 | X531 | X532 | X533 | X534 | X535 | X536 | X537 | X538 | X539 | X540 | X541 | X542 | X543 | X544 | X545 | X546 | X547 | X548 | X549 | X550 | X551 | X552 | X553 | X554 | X555 | X556 | X557 | X558 | X559 | X560 | X561 | X562 | X563 | X564 | X565 | X566 | X567 | X568 | X569 | X570 | X571 | X572 | X573 | X574 | X575 | X576 | X577 | X578 | X579 | X580 | X581 | X582 | X583 | X584 | X585 | X586 | X587 | X588 | X589 | X590 | X591 | X592 | X593 | X594 | X595 | X596 | X597 | X598 | X599 | X600 | X601 | X602 | X603 | X604 | X605 | X606 | X607 | X608 | X609 | X610 | X611 | X612 | X613 | X614 | X615 | X616 | X617 | X618 | X619 | X620 | X621 | X622 | X623 | X624 | X625 | X626 | X627 | X628 | X629 | X630 | X631 | X632 | X633 | X634 | X635 | X636 | X637 | X638 | X639 | X640 | X641 | X642 | X643 | X644 | X645 | X646 | X647 | X648 | X649 | X650 | X651 | X652 | X653 | X654 | X655 | X656 | X657 | X658 | X659 | X660 | X661 | X662 | X663 | X664 | X665 | X666 | X667 | X668 | X669 | X670 | X671 | X672 | X673 | X674 | X675 | X676 | X677 | X678 | X679 | X680 | X681 | X682 | X683 | X684 | X685 | X686 | X687 | X688 | X689 | X690 | X691 | X692 | X693 | X694 | X695 | X696 | X697 | X698 | X699 | X700 | X701 | X702 | X703 | X704 | X705 | X706 | X707 | X708 | X709 | X710 | X711 | X712 | X713 | X714 | X715 | X716 | X717 | X718 | X719 | X720 | X721 | X722 | X723 | X724 | X725 | X726 | X727 | X728 | X729 | X730 | X731 | X732 | X733 | X734 | X735 | X736 | X737 | X738 | X739 | X740 | X741 | X742 | X743 | X744 | X745 | X746 | X747 | X748 | X749 | X750 | X751 | X752 | X753 | X754 | X755 | X756 | X757 | X758 | X759 | X760 | X761 | X762 | X763 | X764 | X765 | X766 | X767 | X768 | X769 | X770 | X771 | X772 | X773 | X774 | X775 | X776 | X777 | X778 | X779 | X780 | X781 | X782 | X783 | X784 | X785 | instance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 18 | 18 | 18 | 126 | 136 | 175 | 26 | 166 | 255 | 247 | 127 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 | 36 | 94 | 154 | 170 | 253 | 253 | 253 | 253 | 253 | 225 | 172 | 253 | 242 | 195 | 64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 49 | 238 | 253 | 253 | 253 | 253 | 253 | 253 | 253 | 253 | 251 | 93 | 82 | 82 | 56 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 219 | 253 | 253 | 253 | 253 | 253 | 198 | 182 | 247 | 241 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 80 | 156 | 107 | 253 | 253 | 205 | 11 | 0 | 43 | 154 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 1 | 154 | 253 | 90 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 139 | 253 | 190 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 190 | 253 | 70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 241 | 225 | 160 | 108 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 81 | 240 | 253 | 253 | 119 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 45 | 186 | 253 | 253 | 150 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 93 | 252 | 253 | 187 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 249 | 253 | 249 | 64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 130 | 183 | 253 | 253 | 207 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 148 | 229 | 253 | 253 | 253 | 250 | 182 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 114 | 221 | 253 | 253 | 253 | 253 | 201 | 78 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 66 | 213 | 253 | 253 | 253 | 253 | 198 | 81 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 171 | 219 | 253 | 253 | 253 | 253 | 195 | 80 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 172 | 226 | 253 | 253 | 253 | 253 | 244 | 133 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 136 | 253 | 253 | 253 | 212 | 135 | 132 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 51 | 159 | 253 | 159 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 238 | 252 | 252 | 252 | 237 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 227 | 253 | 252 | 239 | 233 | 252 | 57 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 60 | 224 | 252 | 253 | 252 | 202 | 84 | 252 | 253 | 122 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 163 | 252 | 252 | 252 | 253 | 252 | 252 | 96 | 189 | 253 | 167 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 51 | 238 | 253 | 253 | 190 | 114 | 253 | 228 | 47 | 79 | 255 | 168 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 238 | 252 | 252 | 179 | 12 | 75 | 121 | 21 | 0 | 0 | 253 | 243 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 165 | 253 | 233 | 208 | 84 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 252 | 165 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 178 | 252 | 240 | 71 | 19 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 252 | 195 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 252 | 252 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 252 | 195 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 198 | 253 | 190 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 255 | 253 | 196 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 76 | 246 | 252 | 112 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 252 | 148 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 252 | 230 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 135 | 253 | 186 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 252 | 223 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 131 | 252 | 225 | 71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 252 | 145 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 165 | 252 | 173 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 86 | 253 | 225 | 0 | 0 | 0 | 0 | 0 | 0 | 114 | 238 | 253 | 162 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 252 | 249 | 146 | 48 | 29 | 85 | 178 | 225 | 253 | 223 | 167 | 56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 252 | 252 | 252 | 229 | 215 | 252 | 252 | 252 | 196 | 130 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 199 | 252 | 252 | 253 | 252 | 252 | 233 | 145 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 128 | 252 | 253 | 252 | 141 | 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 67 | 232 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62 | 81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 120 | 180 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 126 | 163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 153 | 210 | 40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 220 | 163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27 | 254 | 162 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 222 | 163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 183 | 254 | 125 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 245 | 163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 198 | 254 | 56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 120 | 254 | 163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 231 | 254 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 159 | 254 | 120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 163 | 254 | 216 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 159 | 254 | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 86 | 178 | 248 | 254 | 91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 159 | 254 | 85 | 0 | 0 | 0 | 47 | 49 | 116 | 144 | 150 | 241 | 243 | 234 | 179 | 241 | 252 | 40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 150 | 253 | 237 | 207 | 207 | 207 | 253 | 254 | 250 | 240 | 198 | 143 | 91 | 28 | 5 | 233 | 250 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 119 | 177 | 177 | 177 | 177 | 177 | 98 | 56 | 0 | 0 | 0 | 0 | 0 | 102 | 254 | 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 254 | 137 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 254 | 57 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 254 | 57 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 255 | 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 254 | 96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 254 | 153 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 255 | 153 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96 | 254 | 153 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 124 | 253 | 255 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96 | 244 | 251 | 253 | 62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 127 | 251 | 251 | 253 | 62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 68 | 236 | 251 | 211 | 31 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 60 | 228 | 251 | 251 | 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 155 | 253 | 253 | 189 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 253 | 251 | 235 | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 205 | 253 | 251 | 126 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 104 | 251 | 253 | 184 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 80 | 240 | 251 | 193 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 253 | 253 | 253 | 159 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 151 | 251 | 251 | 251 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 221 | 251 | 251 | 172 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 234 | 251 | 251 | 196 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 251 | 251 | 89 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 159 | 255 | 253 | 253 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 228 | 253 | 247 | 140 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 64 | 251 | 253 | 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 64 | 251 | 253 | 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 193 | 253 | 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 148 | 210 | 253 | 253 | 113 | 87 | 148 | 55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 232 | 252 | 253 | 189 | 210 | 252 | 252 | 253 | 168 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 57 | 242 | 252 | 190 | 65 | 5 | 12 | 182 | 252 | 253 | 116 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96 | 252 | 252 | 183 | 14 | 0 | 0 | 92 | 252 | 252 | 225 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 132 | 253 | 252 | 146 | 14 | 0 | 0 | 0 | 215 | 252 | 252 | 79 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 126 | 253 | 247 | 176 | 9 | 0 | 0 | 8 | 78 | 245 | 253 | 129 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 232 | 252 | 176 | 0 | 0 | 0 | 36 | 201 | 252 | 252 | 169 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 252 | 252 | 30 | 22 | 119 | 197 | 241 | 253 | 252 | 251 | 77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 231 | 252 | 253 | 252 | 252 | 252 | 226 | 227 | 252 | 231 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 235 | 253 | 217 | 138 | 42 | 24 | 192 | 252 | 143 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62 | 255 | 253 | 109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 253 | 252 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 253 | 252 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 253 | 252 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 106 | 253 | 252 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 45 | 255 | 253 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 218 | 252 | 56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96 | 252 | 189 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 184 | 252 | 170 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 147 | 252 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 25 | 100 | 122 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 151 | 208 | 252 | 252 | 252 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 152 | 244 | 252 | 253 | 224 | 211 | 252 | 232 | 40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 152 | 239 | 252 | 252 | 252 | 216 | 31 | 37 | 252 | 252 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96 | 252 | 252 | 252 | 252 | 217 | 29 | 0 | 37 | 252 | 252 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 181 | 252 | 252 | 220 | 167 | 30 | 0 | 0 | 77 | 252 | 252 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 128 | 58 | 22 | 0 | 0 | 0 | 0 | 100 | 252 | 252 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 157 | 252 | 252 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 110 | 121 | 122 | 121 | 202 | 252 | 194 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 53 | 179 | 253 | 253 | 255 | 253 | 253 | 228 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 54 | 227 | 252 | 243 | 228 | 170 | 242 | 252 | 252 | 231 | 117 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 78 | 252 | 252 | 125 | 59 | 0 | 18 | 208 | 252 | 252 | 252 | 252 | 87 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 135 | 252 | 252 | 180 | 16 | 0 | 21 | 203 | 253 | 247 | 129 | 173 | 252 | 252 | 184 | 66 | 49 | 49 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 136 | 252 | 241 | 106 | 17 | 0 | 53 | 200 | 252 | 216 | 65 | 0 | 14 | 72 | 163 | 241 | 252 | 252 | 223 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 105 | 252 | 242 | 88 | 18 | 73 | 170 | 244 | 252 | 126 | 29 | 0 | 0 | 0 | 0 | 0 | 89 | 180 | 180 | 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 231 | 252 | 245 | 205 | 216 | 252 | 252 | 252 | 124 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 207 | 252 | 252 | 252 | 252 | 178 | 116 | 36 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 93 | 143 | 121 | 23 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
- Write a function to plot an 28 x 28 image based on a vector, and test it on a row of your data frame.
plot_picture <- function(x) {
x %>%
as.numeric() %>%
matrix(28, 28, byrow = TRUE) %>%
apply(2, rev) %>%
t() %>% image()
}
par(mar = c(0.1,0.1,0.1,0.1))
mnist %>% select(-instance, -label) %>% slice(33) %>% plot_picture()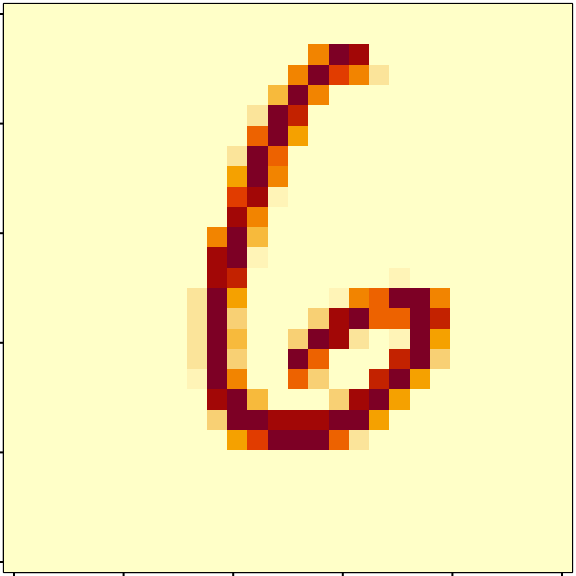
- Make a PCA, choice wether to scale or not.
myPCA <- select(mnist, -instance) %>%
PCA(graph = FALSE, scale.unit = FALSE, quali.sup = 1, ncp = 500)- Make the usual diagnostic plots (screeplot, individual, variable) for the first axes, and comment.
Be careful with the biplot on unscaled PCA.
fviz_eig(myPCA, ncp = 50)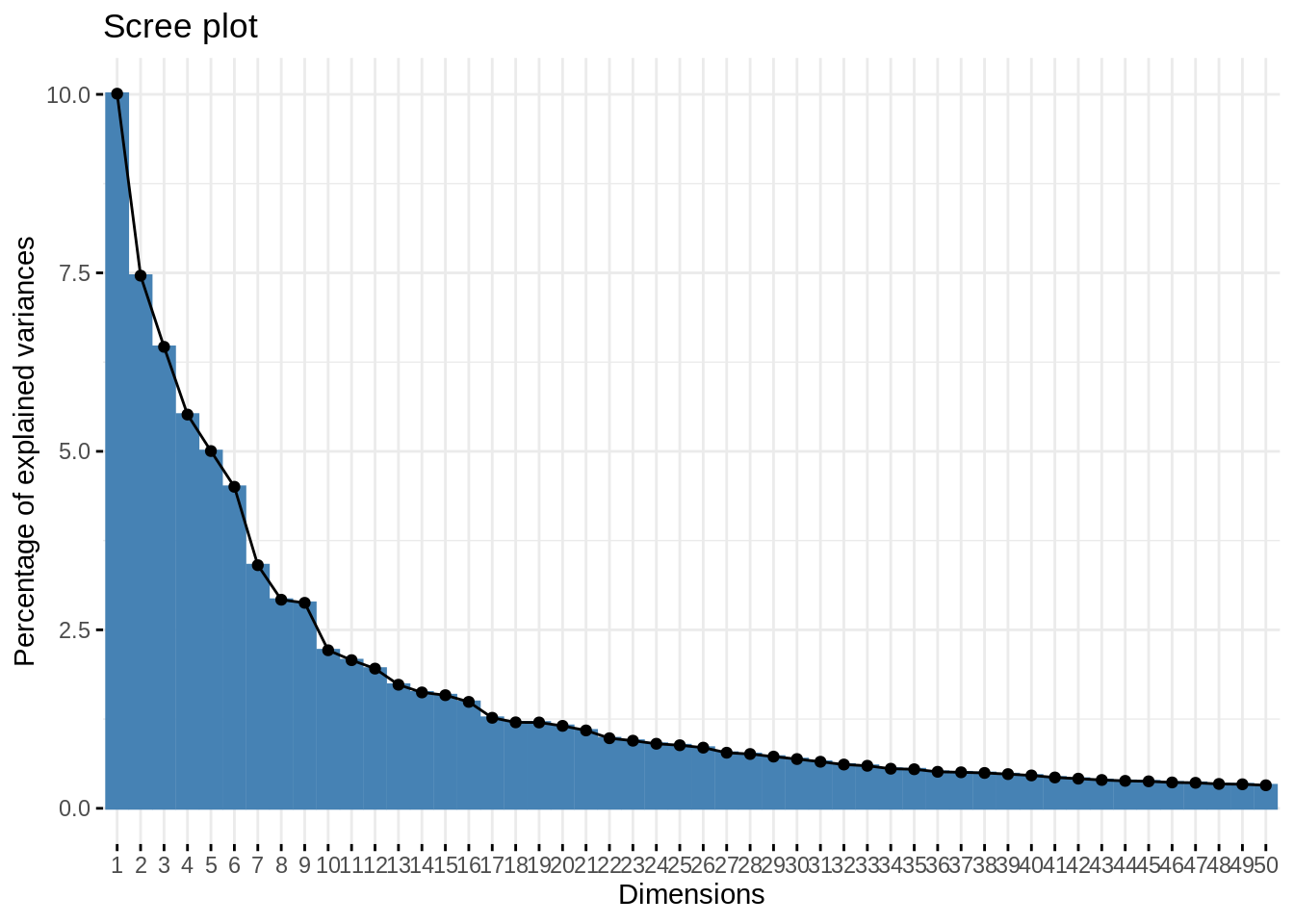
fviz_pca_ind(myPCA, habillage = "label")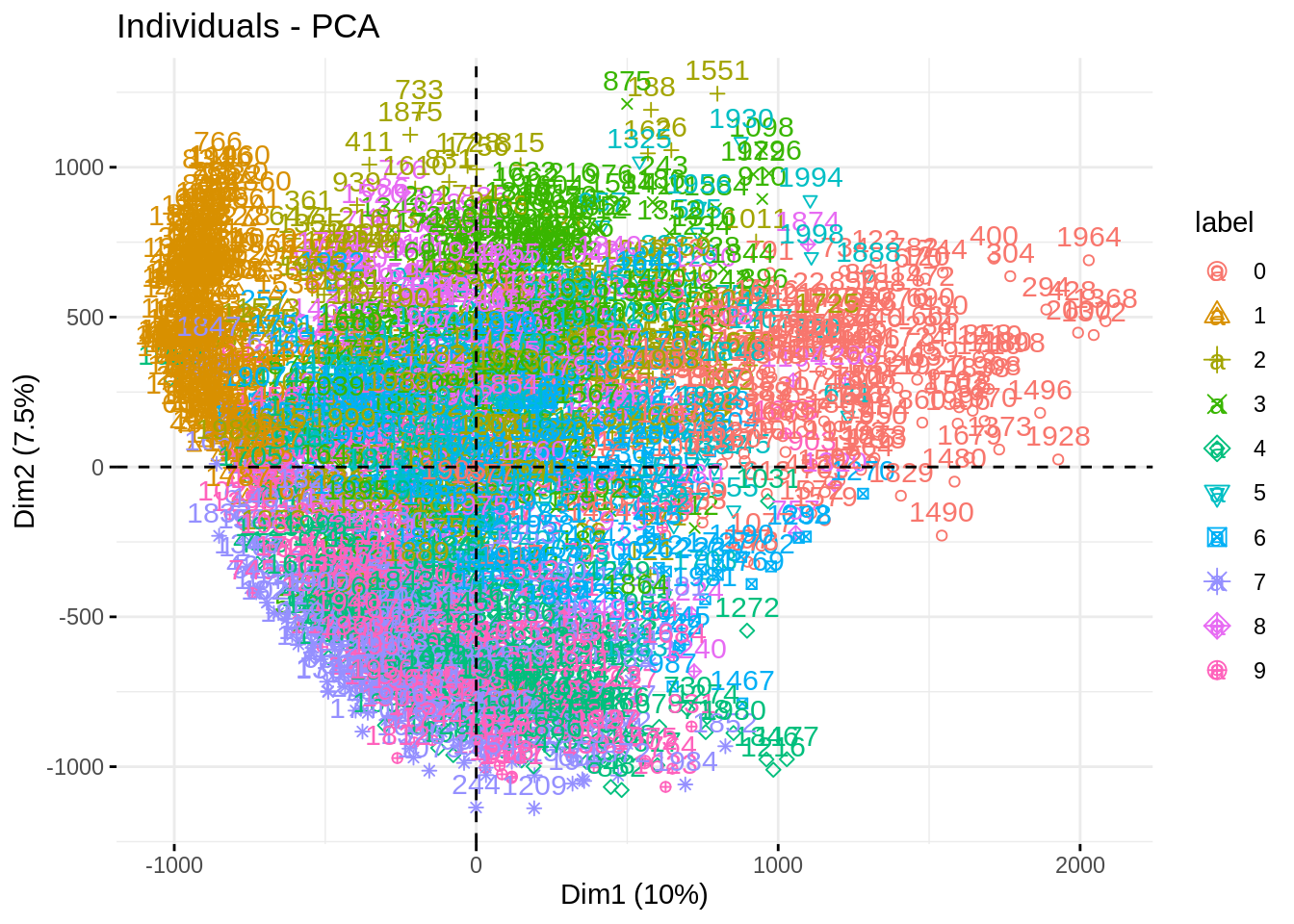
fviz_pca_ind(myPCA, habillage = "label", axes = c(1,3))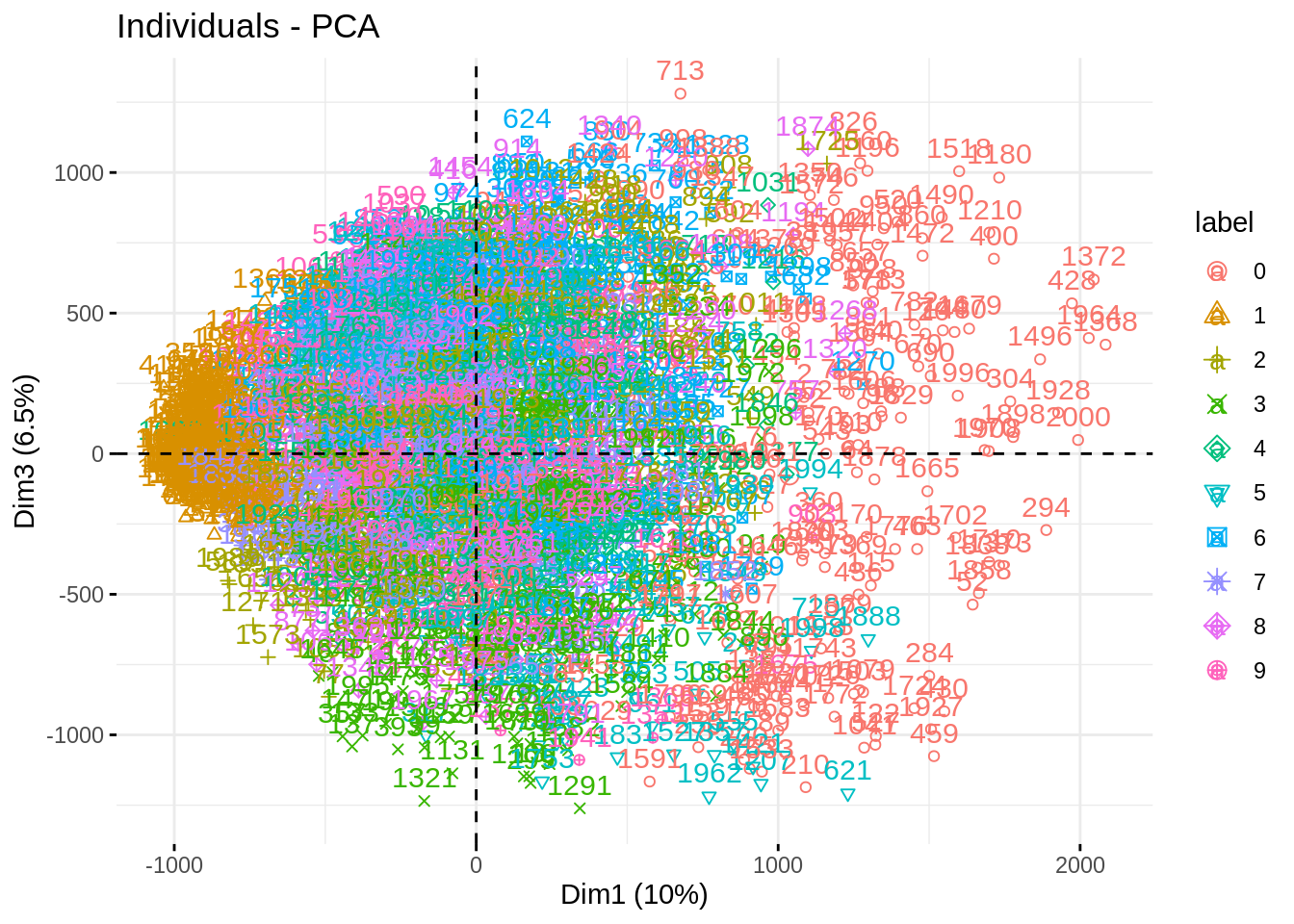
fviz_pca_ind(myPCA, habillage = "label", axes = c(2,3))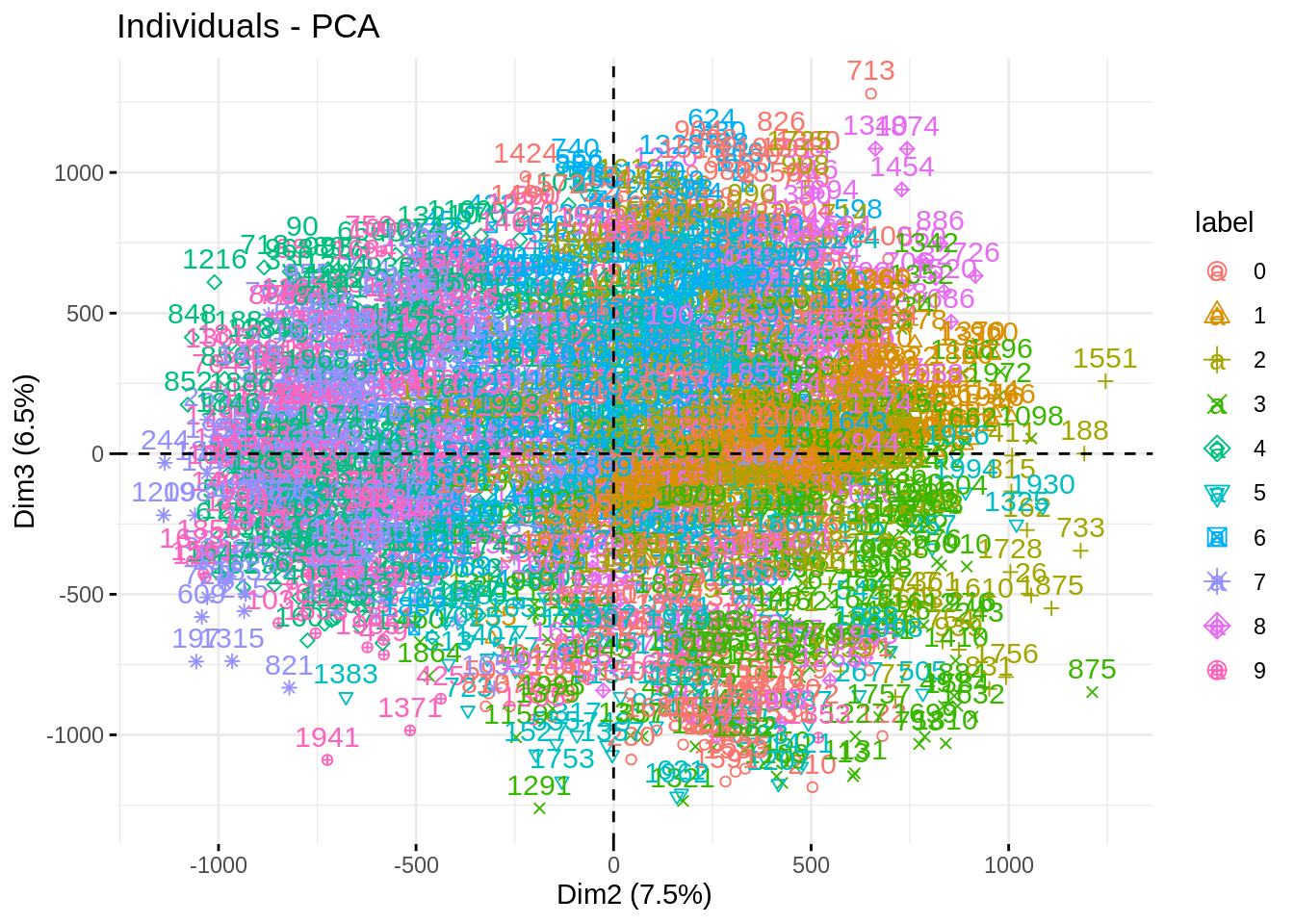
fviz_pca_var(myPCA, col.var = 'cos2', select.var = list(contrib = 30))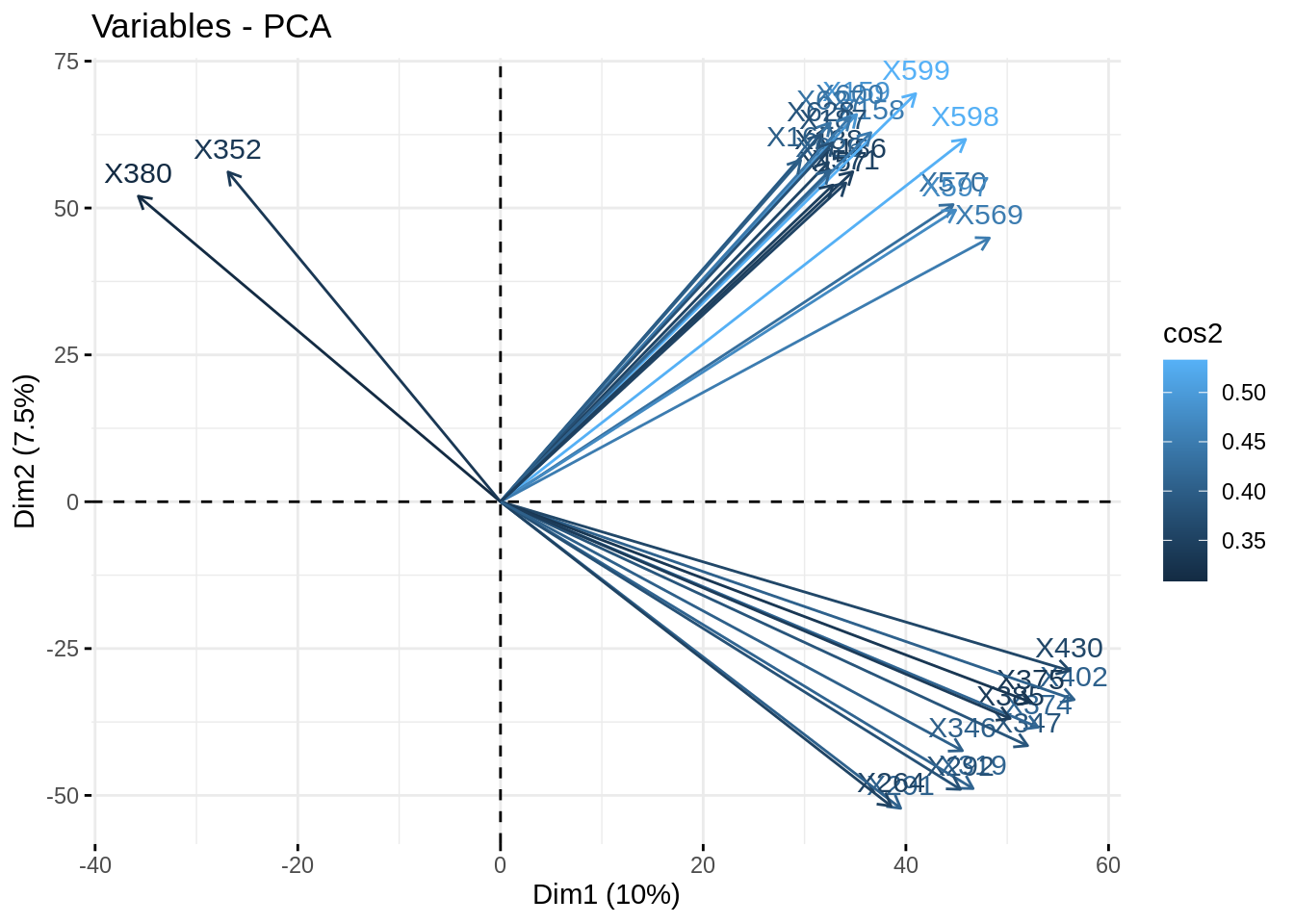
npc <- select(mnist, -instance, -label) %>% replace(is.na(.), 0) %>%
as.matrix() %>% scale(TRUE, FALSE) %>%
estim_ncp(ncp.max = 500, scale = FALSE)
plot(npc$criterion, type = "l", xlab = 'number of component', ylab = 'GCV')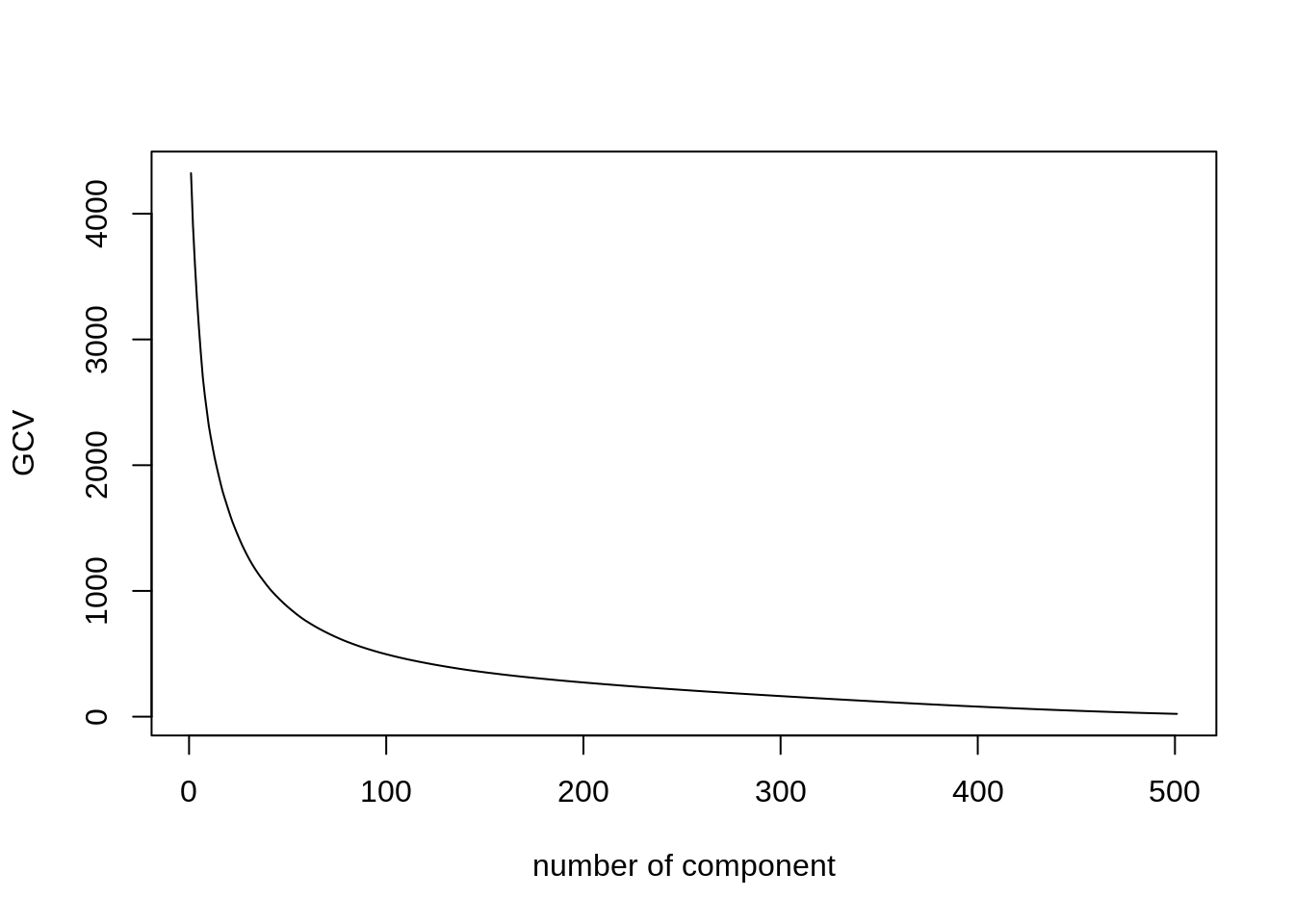
- Write a function to reconstruct an image with two argument: \(i\) (an instance) \(k\) (the number of component used).
## Continuous attributes
X <- select(mnist, -label, -instance) %>% scale(TRUE, FALSE)
## Loadings/rotation matrix
U <- eigen(cov(X))$vectors
## Function for projection
proj_data <- function(k, i) {
X[i, , drop = FALSE] %*% U[, 1:k, drop = FALSE] %*% t(U[, 1:k, drop = FALSE]) %>%
as.numeric()
}- Plot the reconstructed image for various value of \(k\)
Let us pick 9 pictures randomly, and define a sequence for the number of axes used to perform the reconstruction (up to a perfect reconstruction).
nb_samples <- 9
samples <- sample.int(1000, nb_samples)
nb_axis <- c(1, 2, 10, 20, 100, ncol(X))Now here is a solution in ‘base’ R
cases <- expand.grid(nb_axis, samples)
## list of approximated images (stored as vectors)
approx <- mapply(proj_data, cases[, 1], cases[, 2], SIMPLIFY = FALSE)
par(mfrow=c(nb_samples, length(nb_axis)), mar = c(0.1,0.1,0.1,0.1))
silent <- utils::capture.output(lapply(approx, plot_picture))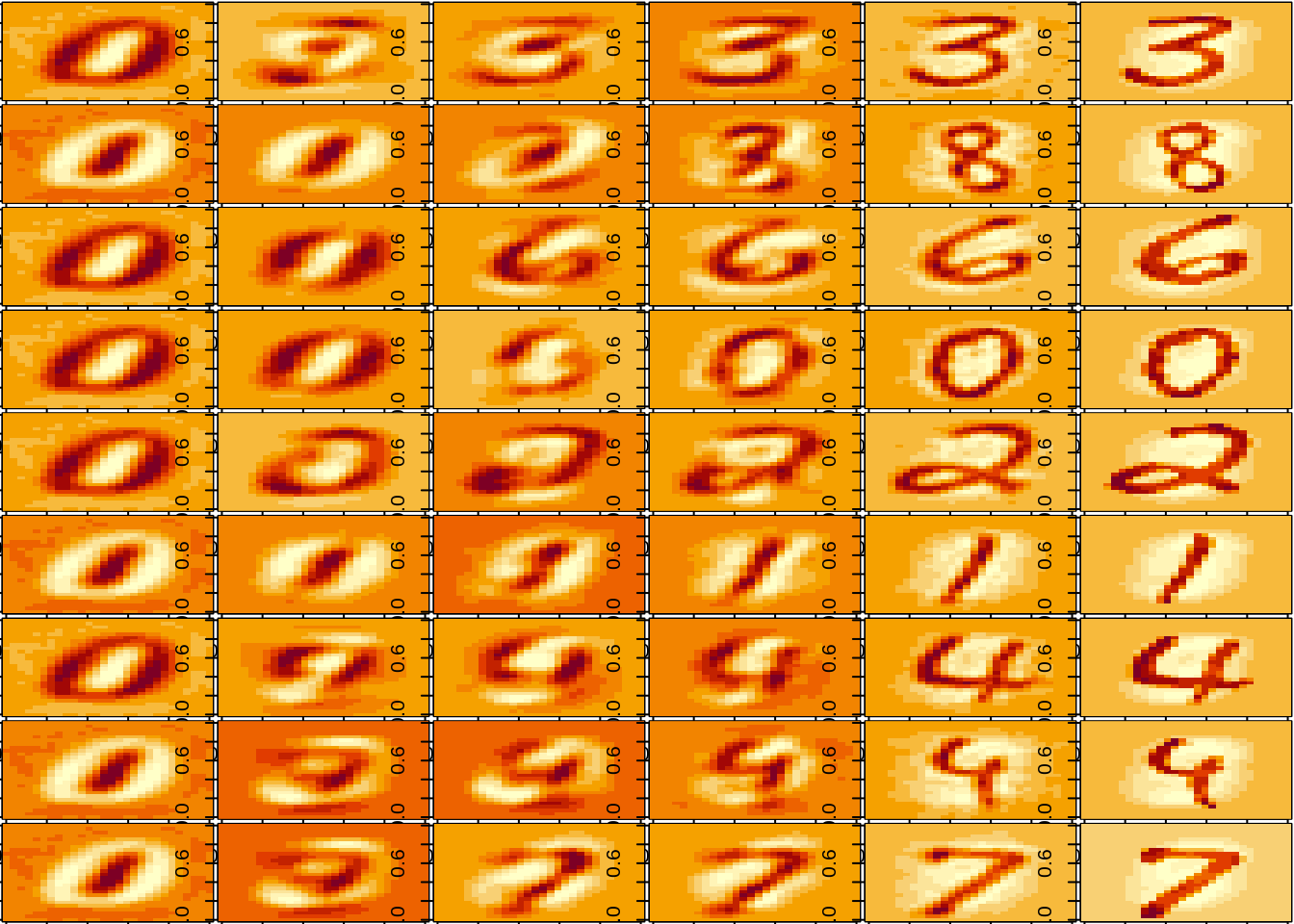
And another solution using {ggplot}.
## fancy ggplot output
labels <- mnist %>% dplyr::filter(instance %in% samples) %>% pull(label)
instances <- mnist %>% dplyr::filter(instance %in% samples) %>% pull(instance)
approx_tibble <- do.call(rbind, approx) %>% as_tibble() %>%
add_column(nb_axis = rep(nb_axis, length(samples)), .before = 1) %>%
add_column(label = rep(labels, each = length(nb_axis)), .before = 1) %>%
add_column(instance = rep(instances, each = length(nb_axis)), .before = 1)## Warning: The `x` argument of `as_tibble.matrix()` must have unique column names if `.name_repair` is omitted as of tibble 2.0.0.
## Using compatibility `.name_repair`.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_warnings()` to see where this warning was generated.approx_tibble %>%
gather(pixel, value, -label, -instance, -nb_axis) %>%
tidyr::extract(pixel, "pixel", "(\\d+)", convert = TRUE) %>%
mutate(pixel = pixel - 2,
x = pixel %% 28,
y = 28 - pixel %/% 28) %>%
ggplot(aes(x, y, fill = value)) +
geom_tile() +
facet_grid(label ~ nb_axis)